The DNA Network |
| Aphid-bacterial symbiosis in more detail, and in the New York Times [The Tree of Life] Posted: 08 Dec 2008 07:41 PM CST Nice little bit in the New York Times tomorrow about aphids and their symbionts. Henry Fountain writes (Observatory - How Tiny Insects, With a Little Help, Survive on Plant Sap - NYTimes.com) about a new article by Angela Douglas, one of the true pioneers of endosymbiont research. In her study she dissects in fine scale detail which essential amino acids are missing from the aphid sap only diet and which ones are made by the symbionts. Interestingly, the research apparently shows that the aphids may have figured out how to make methionine by themselves. I say apparently since I have been unable to track down the paper which I assume is coming out soon. I should note, in one of the symb  ioses like this that I have studied with Nancy Moran we found that there were two symbionts contributing to the nutrition of the host. We found that one of the symbionts was likely making amino acids for the host (an insect called the glassy winged sharpshooter which eats only xylem sap) and the other symbiont was likley making vitamins. Nancy showed later with John McCutcheon that the symbiont that was making vitamins also was predicted to be making methionine for the host. So it seems possible there might be a missing symbiont in the aphid study? Although it would be cool if the aphid has figured out how to make an amino acid most animals are not able to make. ioses like this that I have studied with Nancy Moran we found that there were two symbionts contributing to the nutrition of the host. We found that one of the symbionts was likely making amino acids for the host (an insect called the glassy winged sharpshooter which eats only xylem sap) and the other symbiont was likley making vitamins. Nancy showed later with John McCutcheon that the symbiont that was making vitamins also was predicted to be making methionine for the host. So it seems possible there might be a missing symbiont in the aphid study? Although it would be cool if the aphid has figured out how to make an amino acid most animals are not able to make.Hat tip to Max Lambert for pointing this out. |
| Nils Reinton (SciPhu) relaunches site: BIOpinionated [Think Gene] Posted: 08 Dec 2008 06:23 PM CST Nils Reinton has relaunched his site, SciPhu, as BIOpinionated. Nils is a molecular biology scientist working in medical diagnostics, and his blog is consistently interesting and insightful. Check it out! |
| Tree of Life Gift Recommendation - Climate Kits [The Tree of Life] Posted: 08 Dec 2008 05:33 PM CST Just a quick recommendations for a gift for this holiday seasons that seems cool (metaphorically and literally). It is the climate kit. It comes from a friend of mine from college, Kathy Washienko and this is some of their text: Kits are convenient collections of tools and tips that will help your family and friends reduce your environmental impact. By grouping what you need in one handy package, a kit makes it easy and fun to take energy-saving steps. Each kit is ultimately a gift to our environment, but will also save you money in reduced energy costs.* And every kit comes with our innovative "rebate." Check them out! |
| Save babies, break the law. [Think Gene] Posted: 08 Dec 2008 04:52 PM CST This was such a “good idea” that I had to share. Maybe some radical Ivy grad student can pick up the ball here.
Originally posted as a comment by Andrew Yates on Think Gene using Disqus. |
| The genetic architecture of metabolic traits: a data explosion [Genetic Future] Posted: 08 Dec 2008 03:07 PM CST Nature Genetics has just released six advance online manuscripts on the genetic architecture of complex metabolic traits. The amount of data in the manuscripts is overwhelming, so this post is really just a first impression; I suspect I'll have more to say once I've had time to dig into the juicy marrow of the supplementary data. The general approach of exploring the genetic architecture of quantitative disease-associated traits (often called intermediate phenotypes or endophenotypes) rather than categorical case-control analyses of disease status raises some interesting questions, but I'm going to save those for another time and focus instead on the results. Executive summary For those who are interested in more details, I've tucked them below the fold: Citation: |
| TripAnswers or Twitter? [ScienceRoll] Posted: 08 Dec 2008 03:00 PM CST Twitter is a microblogging tool that allows us to post 140 charachter-long messages and we do have a huge medical community there so it’s quite easy to ask medical questions and get relevant answers from doctors from around the world. TripAnswers is a site where clinicians can ask questions.
Which one would you use to ask questions? Futher reading:
 |
| National physician group MDVIP partners with Navigenics [The Navigator - Navigenics Blog] Posted: 08 Dec 2008 03:00 PM CST Vance Vanier, M.D.
National Physician Group MDVIP Partners with Navigenics to Provide Personal Genetic Tests for Preventive Medicine Practice Boca Raton, FL and Redwood Shores, CA – December 8 2008 – MDVIP, Inc., a leading national network of physicians dedicated to preventive and personalized healthcare, and Navigenics, Inc., a leading personal genomics testing company, today announced a first of its kind collaborative effort to integrate genomic-based preventive healthcare in physician offices. Through this initiative, Navigenics' genomic testing service will be available to MDVIP affiliated physicians to help patients understand their genetic risk factors for disease and work with their doctors to develop individualized prevention plans. Navigenics will provide MDVIP patients and their affiliated physicians with insight into their personal genetic predisposition for developing certain medical conditions where primary or secondary prevention could improve health outcomes. The Navigenics test will identify individuals' genetic markers for developing such conditions as type 2 diabetes, cancer, heart attack, and celiac disease. Working with their personal MDVIP physician and Navigenics' board-certified Genetic Counselors, individuals can chart and implement a personalized wellness course to help decrease their overall risk, delay disease onset or prevent it altogether. "We have for many years been closely watching the field of genomic testing evolve into a tool that can enhance and inform the practice of preventive medicine," said Edward Goldman, M.D., CEO of MDVIP. "We believe that Navigenics' preventive genomics service has the potential to be an innovation that could significantly enhance patient care." |
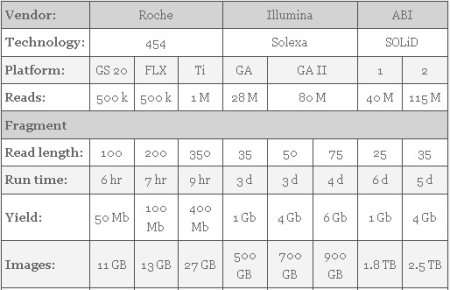
| Next-Generation Sequencing Statistics [ScienceRoll] Posted: 08 Dec 2008 02:52 PM CST Have you ever wondered about the typical run time of a next-generation genome sequencer? PolITiGenomics has the answers: Read more about next-generation sequencing in the Trends in Genetics article: The impact of next-generation sequencing technology on genetics.  |
| Salary prospects for biology majors [Bayblab] Posted: 08 Dec 2008 01:08 PM CST This may or may not surprise you, but who do you think has the highest median starting salary and mid-career salary between those three: History graduate, philosophy graduate, biology graduate? When it comes to salary you're actually better off with an undergrad in history, philosophy, geography, any engineering, or any science other than biology. If it makes you feel better, it still beats drama and is about equivalent to an English major with $38.8K starting and $64.8K mid-career. Why is that? I'm not sure, but It probably is a consequence of supply and demand. Not much demand for biologists, and way too many graduates. |
| Leaf Camouflage Pictures and Videos [Bayblab] Posted: 08 Dec 2008 12:59 PM CST  I ran into a collection of awesome leaf mimicry pictures and videos I origonally ran into on boing boing. Definitely worth checking it out. The blog that is hosting these pictures, The Conservation Report, seems to have an ongoing thing with pictures of impressive animal camouflage. While I would love to get some of the leaf mimicing fish for my aquarium, looks like they'd eat my neon tetras! |
| Finnish population structure won't persist much longer [Genetic Future] Posted: 08 Dec 2008 12:45 PM CST In my previous post on Finnish population clustering I should have emphasised that the map was constructed only from individuals who had both parents from the same geographic/linguistic region; this obviously provides a lot more power to detect a correlation. The close match between genetic and geographic ancestry in these selected individuals indicates that there hasn't been a huge amount of long-range admixture between Finnish populations over quite a long period of time - if there had been, there would be no reason to expect much correlation between the two maps. However, data in the supplementary section of the paper show that there has been substantial mixing over more recent history:
This graph shows the number of individuals from the cohort that had both parents from a single region. You can see immediately that the single largest category is the "mixed" category indicating parentage that crosses region boundaries. It seems likely that many of these individuals come from border areas between the regions (in which case excluding them from the map will have resulted in a rather artificial increase in the separation between clusters), but no doubt there are also many examples of long-range admixture resulting from increases in population mobility during the 20th century. As mobility continues to increase the correlation between geographical location and genetic ancestry will decline rapidly. We really are in a brief and privileged moment in time: a moment when we have the tools to perform genome-scale analyses of ancestry, but before globalisation [and urbanisation] begins to erase information about historical population structure. Chiara Sabatti, Susan K Service, Anna-Liisa Hartikainen, Anneli Pouta, Samuli Ripatti, Jae Brodsky, Chris G Jones, Noah A Zaitlen, Teppo Varilo, Marika Kaakinen, Ulla Sovio, Aimo Ruokonen, Jaana Laitinen, Eveliina Jakkula, Lachlan Coin, Clive Hoggart, Andrew Collins, Hannu Turunen, Stacey Gabriel, Paul Elliot, Mark I McCarthy, Mark J Daly, Marjo-Riitta Järvelin, Nelson B Freimer, Leena Peltonen (2008). Genome-wide association analysis of metabolic traits in a birth cohort from a founder population Nature Genetics DOI: 10.1038/ng.271 Read the comments on this post... |
| Congrats to Pamela Ronald et al. for Award for Flood Resistant Rice [The Tree of Life] Posted: 08 Dec 2008 11:00 AM CST Congrats to Pam Ronald, colleague, Davis faculty member, and fellow science blogger for receiving a USDA Discovery Award for helping develop a flood resistant rice variety. For more on this see
|
| Posted: 08 Dec 2008 10:25 AM CST  Prince Joachim of Denmark and Princess Marie of Denmark along with deCODE scientist Unnur Thorsteinsdottir during an official visit to deCODE laboratories earlier this year. Reykjavik, ICELAND, December 8, 2008 – deCODE genetics (Nasdaq:DCGN) today announced the discovery by an international consortium of scientists from deCODE and major European and US academic institutions of a single letter variation in the human genome (SNP) that is associated with increased fasting glucose levels and risk of type 2 diabetes (T2D). deCODE will employ its CLIA-registered genotyping laboratory and existing testing platform to swiftly integrate the finding into its deCODEme™ personal genome scan, and to assess the addition of this new variant to the company's deCODE T2™ reference laboratory test for assessing individual risk of type 2 diabetes. "This finding is another step towards rounding out our understanding of the genetic factors that underpin glucose regulation and risk of type 2 diabetes. This variant does not confer sufficient risk to be of clinical utility on its own. But when measured in addition to our TCF7L2 variant that is the anchor of the deCODE T2™ test, it may, like other common variants conferring modest risk, enable the test to capture an even larger proportion of inherited risk. We are currently evaluating its integration into deCODE T2™, because understanding genetic risk of T2D enables individuals and their physicians to focus, personalize and improve prevention. In the meantime, we will be enabling our deCODEme subscribers to check their profiles for this new variant, keeping them at the cutting edge of human genetics" said Kari Stefansson, CEO of deCODE. Type 2 diabetes: A major public health problem |
| 23andMe offers family discount, just in time for Christmas [Genetic Future] Posted: 08 Dec 2008 09:45 AM CST Personal genomics company 23andMe is now offering a discount of $200 for customers who buy three or more kits before December 31st. In a press release the company explains the reasoning behind the price cut: By offering this discount, 23andMe hopes to encourage families, in particular, to explore the unique features of the 23andMe Personal Genome Service™ that are of special interest to people who are related. These features allow family members to learn how genetically similar they are and how genes were passed down from grandparents to grandchildren. Still unconvinced? Just imagine the heart-warming sound of your entire family spitting into plastic tubes on Christmas morning. Now that is the sound of the 21st century Christmas...
|
| Genetics and geography in Finland [Genetic Future] Posted: 08 Dec 2008 08:35 AM CST Here's a figure from a brand new paper on the genetics of metabolic traits in a large Finnish cohort (which I'll be posting about in more detail shortly): The Finns are an interesting group, being clear genetic outliers relative to most of the rest of Europe. No doubt Razib will have more to say about this map given his notorious attitudes towards the Finnish people. Added in edit: It's worth emphasising that the individuals plotted here were only those who had both parents from the same region; see my follow-up post. Chiara Sabatti, Susan K Service, Anna-Liisa Hartikainen, Anneli Pouta, Samuli Ripatti, Jae Brodsky, Chris G Jones, Noah A Zaitlen, Teppo Varilo, Marika Kaakinen, Ulla Sovio, Aimo Ruokonen, Jaana Laitinen, Eveliina Jakkula, Lachlan Coin, Clive Hoggart, Andrew Collins, Hannu Turunen, Stacey Gabriel, Paul Elliot, Mark I McCarthy, Mark J Daly, Marjo-Riitta Järvelin, Nelson B Freimer, Leena Peltonen (2008). Genome-wide association analysis of metabolic traits in a birth cohort from a founder population Nature Genetics DOI: 10.1038/ng.271 Read the comments on this post... |
| I'll be hosting the 4th Molecular and Cell Biology Carnival on December 14th [The Daily Transcript] Posted: 08 Dec 2008 08:18 AM CST Send in your entries before the weekend using this form or by emailing me directly. You can visit the MCB Carnival homepage by clicking on the badge/button: |
| Advice for doctors on dealing with personal genomics customers [Genetic Future] Posted: 08 Dec 2008 06:52 AM CST Think Gene's Andrew Yates has posted generic responses for medical professionals to use when dealing with patients who come armed with their results from 23andMe or Navigenics. They're probably quite useful little tools for busy doctors without the time to brush up on the field of personal genomics, but - seeing as this is Andrew Yates - they're also a dig at the careful "medicine but not medicine" stance of personal genomics companies. An excerpt: Thus, applying 23andMe to your health care would be a violation of the 23andMe terms of service and, as stated, it "cannot be relied upon at this point for diagnostic purposes." We think 23andMe is a great educational tool, and we are excited about its future potential, but we cannot use the test results to provide any medical services. Further, you consented to "not change your health behaviors on the basis of [23andMe]," so for us to counsel otherwise would be unethical.Read the comments on this post... |
| Calling fellow bloggers! [Genomicron] Posted: 08 Dec 2008 06:40 AM CST Calling fellow bio bloggers -- help get the word out for the special issue of Evolution: Education and Outreach all about eye evolution. The content is free to access online and the authors include many of the world's top eye evolution researchers. A handy table of contents with links is provided below for easy copy and paste maneuvers. Evolution: Education and Outreach Volume 1 Issue 4 Editorial 351. Editorial by Gregory Eldredge and Niles Eldredge (PDF) 352-354. Introduction by T. Ryan Gregory (PDF) 355-357. Casting an Eye on Complexity by Niles Eldredge (PDF) Original science / evolution reviews 358-389. The Evolution of Complex Organs by T. Ryan Gregory (PDF) (Blog: Genomicron) 390-402. Opening the "Black Box": The Genetic and Biochemical Basis of Eye Evolution by Todd H. Oakley and M. Sabrina Pankey (PDF) (Blog: Evolutionary Novelties) 403-414. A Genetic Perspective on Eye Evolution: Gene Sharing, Convergence and Parallelism by Joram Piatigorsky (PDF) 415-426. The Origin of the Vertebrate Eye by Trevor D. Lamb, Edward N. Pugh, Jr., and Shaun P. Collin (PDF) 427-438. Early Evolution of the Vertebrate Eye—Fossil Evidence by Gavin C. Young (PDF) 439-447. Charting Evolution's Trajectory: Using Molluscan Eye Diversity to Understand Parallel and Convergent Evolution by Jeanne M. Serb and Douglas J. Eernisse (PDF) 448-462. Evolution of Insect Eyes: Tales of Ancient Heritage, Deconstruction, Reconstruction, Remodeling, and Recycling by Elke Buschbeck and Markus Friedrich (PDF) 463-475. Exceptional Variation on a Common Theme: The Evolution of Crustacean Compound Eyes by Thomas W. Cronin and Megan L. Porter (PDF) 476-486. The Causes and Consequences of Color Vision by Ellen J. Gerl and Molly R. Morris (PDF) 487-492. The Evolution of Extraordinary Eyes: The Cases of Flatfishes and Stalk-eyed Flies by Carl Zimmer (PDF) (Blog: The Loom) 493-497. Suboptimal Optics: Vision Problems as Scars of Evolutionary History by Steven Novella (PDF) (Blog: NeuroLogica) Curriculum articles 498-504. Bringing Homologies Into Focus by Anastasia Thanukos (PDF) (Website: Understanding Evolution) 505-508. Misconceptions About the Evolution of Complexity by Andrew J. Petto and Louise S. Mead (PDF) (Website: NCSE) 509-516. Losing Sight of Regressive Evolution by Monika Espinasa and Luis Espinasa (PDF) Book reviews 548-551. Jay Hosler, An Evolutionary Novelty: Optical Allusions by Todd H. Oakley (PDF) |
| Posted: 08 Dec 2008 06:00 AM CST I was planning to write a long article on this recent paper in PLoS Genetics, but p-ter at Gene Expression and G at Popgen ramblings have both covered the central message very well. So if you haven't read those articles, already, go and do so now - when you come back, I want to talk about the potentially worrying implications of this paper for the future of personal genomics. Read the rest of this post... | Read the comments on this post... |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |





No comments:
Post a Comment