The DNA Network |
| First reading on Tomorrow's Table [Tomorrow's Table] Posted: 25 Apr 2008 06:59 PM CDT Last night Raoul and traveled to Mrs. Dalloway's bookstore in Berkeley to discuss our book "Tomorrow's Table". Here are some questions/comments from the audience and our answers: Beth Greenfield, stellar yoga teacher in Berkeley: " It does seem like a very complicated subject when you consider everyone's opinions and prejudices. How do you prove that GE produce is safe to consume and grow? And would diehard anti-GE people even listen? You made a good case when you compared it to GM produce and that it is considered okay. But I guess people still think "don't fool with Mother Nature". Unknown citizen "How can you prove the nutritional content of GE food is not damaged?" Pam: "Of all the concerns about GE food, this question of risk presents the biggest worry for the most people. The U.S. National Academy of Science addressed a broader question by asking, 'Is the process of adding genes to our food by genetic engineering any more risky than adding genes by traditional breeding?' The answer is no. Virtually everything we eat has been genetically modified in someway, and virtually every food we eat poses some kind of risk, albeit a very, very small risk. The NAS committee determined that both the process of GE and conventionalbreeding pose similar risks of unintended consequences." Jon Agee, author of children's picture books, husband of Audrey and idol of our children. "I don't think you reached the woman that asked about nutrition. She left early and did not buy a book". Old friend of Raoul's, Tom Schwartz, Berkeley: "I heard that transgenes were found in traditional corn varieties in Mexico, is this a problem?" Pam: "In a study of corn landraces in Northern Oaxaca, Ignacio Chapela, a professor at the University of California, Berkeley, published a paper in Nature providing evidence for the presence of transgenic DNA in these landraces. The published results ignited an explosion of worldwide publicity because transgenic corn had never been approved for cultivation in Mexico and there was concern that the presence of transgenes might compromise the genetic diversity of these landraces. Although the results presented in the initial publication were widely disputed and then refuted by a larger peer-reviewed study, the paper prompted an important debate over possible biological, economic, and cultural implications of gene flow. These issues are increasingly important because Mexican corn growers want to use GE to improve productivity and poor consumers rely on this staple. Each crop GE or conventionally bred variety needs to be evaluated on a case-by-case basis as to whether gene flow will create a problem in a particular environment. In California's central valley it is not such an issue, because most of the crops grown here (GE and not GE) do not cross-pollinate native species nearby. It is as if California were a large, oval-shaped, flat-bottomed platter with steep, slippery sides holding all the domesticated crop plants at the bottom." Rick Ronald, favorite younger brother: "You should cut the talk in half and leave more time for questions- that part was hot." Unknown "I hear that the farmer Percy Schmeiser was sued by Monsanto and it was nothis fault. It sounds like he had a terrible time. Pam, passing on the question to an audience member. "Peggy (UCB professor and creator of ucbiotech.org), would you be willing to comment?" Peggy: "I have heard Percy talk and read some of the court documents. The court found that Schmeiser had either known or ought to have known that he had planted Roundup Ready canola in 1998. The court also found that by selling the seed harvested in 1998 Percy further infringed Monsanto's patent." Mateo Burtch, tech writer and funny guy "You spelled my name wrong in the acknowledgements" Mrs. Dalloway's staffer 30 minutes after closing time "Would you please leave now?" |
| Disclosing Risk: Good Communication or "Doctor-Knows-Best"? [PredictER Blog] Posted: 25 Apr 2008 03:58 PM CDT A newly published paper from PredictER's Peter H. Schwartz and Eric M. Meslin, examines the challenges of balancing beneficence and the respect for autonomy in preventive and predictive medicine. In "The ethics of information: absolute risk reduction and patient understanding of screening" (J Gen Intern Med. 2008 Apr 18; [Epub ahead of print] | PMID: 18421509) the authors question whether providing absolute probabilities of risk based, for example, on genetic screening for breast cancer, is always in the best interest of the patient's health. While many argue the respect for the patient's autonomy demands that risk is communicated numerically or graphically, Schwartz and Meslin argue that the disclosures should be made "in the light of careful consideration of patient understanding and possible impacts on uptake and well-being". |
| Happy DNA Day!!! [The Gene Sherpa: Personalized Medicine and You] Posted: 25 Apr 2008 03:24 PM CDT |
| SimpleGenetics(TM) for DNA day [www.cancer-genetics.com] Posted: 25 Apr 2008 03:13 PM CDT Happy DNA day! This year in Europe we celebrate it for the first time officially. What a coincidence - yeasterday I’ve received gene T-shirt with mapped 1st chromosome from the magazine Science - wearing it all this day
On this occasion I would like to introduce you to SimpleGenetics(TM) genetic test reviews - it could be an independent wiki-based resource dedicated to the provision of reviews and unbiased information about commonly available genetic tests, its clinical validity and utility. The project aims to improve the quality and accessibility of information regarding genetic tests and help to make informed choice decision for physicians and/or patients. The information would be reviewed and edited by registered clinicians or scientists working in clinical genetic settings. There is a preference for objective, scientific evidence-based, comprehensive reviews of clinical evidence and appreciate contribution of both genetic testing companies, clinicians and users to satisfy these goals. I made this draft in February, and there is some information about Mammaprint available already. Let me know what you think about it and future involvement/contribution/etc. possibility. Check out at SimpleGenetics.com  |
| Health World Web: A Health 2.0 Platform [ScienceRoll] Posted: 25 Apr 2008 02:21 PM CDT Health World Web, Inc., a Jacksonville, FL company, that tries to create a new, more interactive platform for patients and medical professionals as well. It offers emotional support, non-medical advice; ratings, reviews and recommendations for local doctors, dentists and chiropractors. The database now consists of more than a hundred patient communities and nearly 1.5 million doctors. An excerpt from the mission statement:
And here is a video describing the main goals and features of the site. If you mix social networking and doctor search with a twist, it is called HealthWorldWeb.com More at Health World Web… This is a cross-post from Medgadget.  |
| Personalized Medicine: Real Clinical Examples! [ScienceRoll] Posted: 25 Apr 2008 02:00 PM CDT
References:
 |
| Celebrate DNA Day! [DNA Direct Talk] Posted: 25 Apr 2008 01:02 PM CDT Happy DNA Day, everybody! Today is great opportunity to celebrate DNA — whether you’re a student, an expert, a science lover or a novice. So put on your DNA t-shirts, get out there and hug a helix. (I’d love be a grown-up kid at Dr. Barry Star’s events at the Tech Museum of Innovation today, [...] |
| Phylogeny Friday -- 25 April 2008 [evolgen] Posted: 25 Apr 2008 10:00 AM CDT Duh! That's Obvious, Edition Take a look at this mastodon skeleton: Does it look like anything you recognize? Perhaps a large terrestrial mammal with big tusks. If you said "elephant" you win. The prize: nothing. That is half of the conclusion from a recent paper in Science (doi:10.1126/science.1154284). Really. The other half: birds and dinosaurs are pretty closely related. Or, more specifically, birds and Tyrannosaurus rex -- THE COOLEST MOST AWESOMEST OF ALL DINOSAURS EVER!! -- are closely related. And, for this, they get a Science paper. Now, the way they did this is pretty damn cool: they sequenced proteins from T. rex bones. But that was reported last year (doi:10.1126/science.1137614) in the paper where they screwed up the species name in the title (they got it right this time around). Anyway, some of the same people took those sequences and, along with some other sequences that they mined from various databases, constructed a phylogenetic tree of vertebrates. Read the rest of this post... | Read the comments on this post... |
| Managing Digital Gene Expression Workflows with FinchLab [FinchTalk] Posted: 25 Apr 2008 08:25 AM CDT |
| Posted: 25 Apr 2008 07:04 AM CDT Be a nerd and celebrate National DNA Day with us! DNA Diagnostics Center has two DNA Day eCards–Happy DNA Day Helix Flower and Do The Twist on DNA Day–in honor of the event. (HT: DNA Network member The DNA Testing Blog’s Top Ten Things to Do on DNA Day!)
The National Human Genome Research Institute has more activities planned for National DNA Day including a live online chatroom and DNA Day speaker presentations. |
| Birds and dinosaurs, collagen, and Huxley. [Genomicron] Posted: 25 Apr 2008 07:01 AM CDT I have already blogged about this topic in a short post [Yet another link between dinosaurs and birds], so I will just point out the existence of another paper along these lines using phylogenetic analyses of collagen amino acid sequences recovered from T. rex fossils to demonstrate a close affinity between theropod dinosaurs and birds (Organ et al. 2008), a link first proposed by T.H. Huxley in 1868. This newest analysis was led by Chris Organ, who also did the dinosaur cell/genome size estimation study (Organ et al. 2007; Zimmer 2007). They also looked at mastodons. The data from mastodons and dinosaur aren't new (Asara et al. 2007; Schweitzer et al. 2007) -- the main update is the inclusion extant species and of a phylogenetic component.  There are plenty of news stories about it, including the following. Molecular Analysis Confirms Tyrannosaurus Rex's Evolutionary Link To Birds (ScienceDaily) From T. Rex to Chicken: The Dino-Bird Connection(Discovery News) T. rex confirmed as great granddaddy of all birds (New Scientist) Phylogenetic Tree: Dinosaurs, Alligators And ... Ostriches? (Scientific Blogging) Gunk in T. Rex Fossil Confirms Dino-Bird Lineage (LiveScience) Finally, a reminder about how species names should be listed. _______ References Asara, J.M., M.H. Schweitzer, L.M. Freimark, M. Phillips, and L.C. Cantley. 2007. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science 316: 280-285. Huxley, T.H. 1868. On the animals which are most nearly intermediate between birds and the reptiles. Annals and Magazine of Natural History, Series 4, 2: 66-75. Organ, C.L., A.M. Shedlock, A. Meade, M. Pagel, and S.V. Edwards. 2007. Origin of avian genome size and structure in non-avian dinosaurs. Nature 446: 180-184. Organ, C.L., M.H. Schweitzer, W. Zheng, L.M. Freimark, L.C. Cantley, and J.M. Asara. 2008. Molecular phylogenetics of mastodon and Tyrannosaurus rex. Science 320: 499. Zimmer, C. 2007. Jurassic genome. Science 315: 1358-1359. |
| Biomonitors [Sciencebase Science Blog] Posted: 25 Apr 2008 07:00 AM CDT
Keeping a weather eye on atmospheric pollution is a large-scale, costly and time-consuming activity. However, there just happens to be a vast network of self-contained, self-powered units around the globe that can respond to the presence of toxins, radioactive species, atmospheric particulates and other materials in the environment and could be used to build up a local, national or international picture of environmental conditions - the world’s plants, mosses, and lichens. In a forthcoming special issue of the International Journal of Environment and Pollution (2008, Volume 32, Issue 4), researchers from various fields explain how living organisms can be used to track the dispersal of atmospheric pollutants, particulates, and trace elements. They also explain how plants and other so-called biomonitors have been validated across the globe. Writing in an editorial for the IJEP special issue chemist Borut Smodiš, a senior research associate at the Jožef Stefan Institute, in Ljubljana, Slovenia, explains how biomonitoring can be used in environments where a technological approach to monitoring is not only difficult and costly but may be impossible. “Biomonitoring allows continuous observation of an area with the help of bioindicators, an organism (or part of it) that reveals the presence of a substance in its surroundings with observable and measurable changes (e.g. accumulation of pollutants), which can be distinguished from the effects of natural stress.” Smodiš points to numerous other advantages of biomonitoring: “Simple and inexpensive sampling procedures allow a very large number of sites to be included in the same survey, permitting detailed geographical patterns to be drawn. Biomonitoring can be an effective tool for pollutant mapping and trend monitoring in real time and retrospective analysis,” he says. While any organism might be used as a biomonitoring agent, Smodiš points out that mosses and lichens, which lack root systems, are dependent on surface absorption of nutrients. This means that they accumulate particulates and dissolved chemical species from their surroundings rather than from the soil and so could be more appropriate biomonitors for atmospheric pollutants. In 1998, the International Atomic Energy Agency part of the United Nations, started a Coordinated Research Project on biomonitoring. Several papers in the special issue of IJEP detail methodologies, case studies and other aspects of various projects within this initiative and point to future avenues that might be explored.
In the paper “Atmospheric dispersion of pollutants in Sado estuary (Portugal) using biomonitors”, Maria do Carmo Freitas of the Instituto Tecnológico e Nuclear Reactor, in Sacavém, Portugal, and colleagues used instrumental neutron activation analysis (INAA) and proton-induced X-ray emission (PIXE) to investigate pollutant levels in epiphytic lichens. They found that temperature and humidity had a more prominent effect on pollutant accumulation than wind direction or rainfall levels, which could affect the interpretation of other biomonitoring results. Ni Bangfa of the China Institute of Atomic Energy, Beijing, and colleagues in their paper “Study on air pollution in Beijing’s major industrial areas using multielements in biomonitors and NAA techniques” used NAA to analyze three types of plant leaves from Chinese white poplar, arborvitae, and pine needles. They found that northeast Beijing is a clean area while southwest is relatively polluted. In “Biomonitoring in the forest zone of Ghana” B.J.B. Nyarko of the Ghana Atomic Energy Commission and colleagues studied the distribution of heavy metals in agricultural, industrial and mining areas in the first survey of its kind in Ghana using lichens as biomonitors. They found that the area around gold mining regions were most heavily polluted, with arsenic, antimony, and chromium while industrial sites had raised levels of aluminum, iron, and titanium. Farming regions were much less affected by heavy metal pollutants, as one might expect. H.Th. Wolterbeek of the Delft University of Technology, Delft, the Netherlands in ” Large-scale biomonitoring of trace element air pollution: local variance, data comparability and its relationships to human health” used biomonitoring data to determine air concentrations and metal deposition and discussed how such studies might be used in the future to correlate pollution with human health issues. Other researchers including Bernd Markert of International Graduate School Zittau, Zittau, Germany, Eiliv Steinnes of the Norwegian University of Science and Technology, in Trondheim, and their respective teams also further validated the potential of biomonitoring approaches to pollution.
While biomonitoring techniques are improving rapidly and researchers are quickly validating results at the local level, Smodiš points out that there is no single species that could be used on the global scale. Moreover, different weather conditions around the globe mean that techniques are not necessarily comparable. With that in mind, environmental sensor manufacturers may rest assured that there is still a market for their instrumentation despite the best efforts of the mosses and lichens. |
| Around the Blogs [Bitesize Bio] Posted: 25 Apr 2008 04:06 AM CDT Let’s see what’s been going on around the blogs, shall we? From one whose dream of tenure track just came true: And from those newly in a tenure track position, a series of posts discussed - The Public and Science The Personal Genome discussion - Sandra covers a panel discussion with some big names at the University of Washington. Genes and the Environment - Some thoughts on the eternal “nature vs. nurture” debate, in an era of genomics and PLoS. Science Itself (titles say it all) How Can Chromosome Numbers Change |
| Books About DNA: The Century of the Gene by Evelyn Fox Keller [Eye on DNA] Posted: 25 Apr 2008 03:07 AM CDT
In a CBC Radio interview, Dr. Evelyn Fox Keller talks more about genes and public perception. (HT: Women in Science) For more discussion on what is a gene, see this Genome Research article - What is a gene, post-ENCODE? History and updated definition.
DNA Network member Sandra Porter at Discovering Biology in a Digital World gave her definition of a gene last year. |
| The dawn of human matrilineal diversity [HENRY » genetics] Posted: 25 Apr 2008 01:44 AM CDT Out in the AJHG, is the next in the National Genographic line of studies. This one, by Behar et al. assesses 624 mitochondrial genomes from Khoisan hunter gatherers in Africa (doi). The abstract says:
ScienceDaily has some more breathless coverage about this - quoth Spencer Wells: Tiny bands of early humans, forced apart by harsh environmental conditions, coming back from the brink to reunite and populate the world. Truly an epic drama, written in our DNA.. Epic, indeed. |
| New technology for boosting vaccine efficiency [Think Gene] Posted: 25 Apr 2008 01:22 AM CDT One of the most pressing biomedical issues is the development of techniques that increase the efficiency of vaccines. In a paper published on April 24, 2008 in the journal Vaccine, a Massachusetts's biotechnology company, Cure Lab, Inc. has proposed a new technology for anti-viral vaccination. This technology consists of two major elements. First, each vaccine [...] |
| Two suppressor molecules affect 70 genes in leukemia [Think Gene] Posted: 25 Apr 2008 01:20 AM CDT By restoring two small molecules that are often lost in chronic leukemia, researchers were able to block tumor growth in an animal model. The research, using human chronic lymphocytic leukemia (CLL) cells, also showed that loss of the two molecules affects 70 genes, most of which are involved in critical functions such as cell growth, death, [...] |
| Genetic sequencing of protein from T. rex bone confirms dinosaurs’ link to birds [Think Gene] Posted: 25 Apr 2008 01:19 AM CDT Scientists have put more meat on the theory that dinosaurs’ closest living relatives are modern-day birds. Molecular analysis, or genetic sequencing, of a 68-million-year-old Tyrannosaurus rex protein from the dinosaur’s femur confirms that T. rex shares a common ancestry with chickens, ostriches, and to a lesser extent, alligators. The dinosaur protein was wrested from a fossil T. [...] |
| World Premiere of Next Generation Sequencing solution from CLC bio [Next Generation Sequencing] Posted: 25 Apr 2008 01:00 AM CDT Boston, USA — April 25, 2008 — On Monday, April 28, CLC bio will officially unveil their new Next Generation Sequencing solution, CLC Genomics Workbench, the first comprehensive analysis package which can analyze and visualize data from all the major Next Generation Sequencing (NGS) platforms, such as SOLiD from Applied Biosystems, 454 GS flx from [...] |
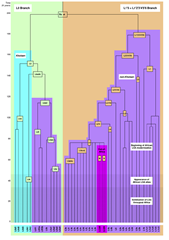
| Human mtDNA Diversity Before Migration Out of Africa [The Genetic Genealogist] Posted: 25 Apr 2008 12:00 AM CDT Two mtDNA Branches The human mtDNA tree has two main branches, the L0 branch which includes individuals concentrated in southern and eastern Africa, and the L1′2′3′4′5′6′ branch (aka the L1′5 branch), which includes the entire remainder of humanity including non-Africans (see the figure to the left). Based upon the analysis of the 624 genomes, the researchers hypothesized that the L0 and L1′5 branches diverged into two small populations around 140,000 to 210,000 years ago, with one group settling in eastern Africa (the L1′5 branch) and the other settling in southern Africa (the L0 branch). Interestingly, the results also suggest that there was little to no intermingling of these branches for the next 50,000 to 100,000 years! The L0 branch comprises 60% of the Khoisan people (two ethnic groups named the Khoi and the San) of Southern Africa. The L1′5 branch comprises all other branches of the mtDNA tree, which includes the N and M matrilines that eventually spread out from Africa. Population Bottleneck The results of the study also suggest that humanity was once comprised of a relatively small number of individuals (as few as 2,000, according to another study cited by the researchers). This was suggested because there are very few matrilineal lineages present today that split during the first 100,000 years of our species’ history, likely because they they died out or never developed in the first place. If there had been many more individuals alive at that time (with descendants alive today), scientists would expect to see more different types of lineages in Africa. This is what happened during the second 100,000 years of human history, with as many as 40 different matrilines at the time that humans left Africa to spread to the remainder of the world. As many of us know now, there is a multitude of current matrilines because of our enormous population explosion in the past few thousand years. Note, however, that this hypothesis may change if researchers suddenly discover large amounts of new matrilines present in Africa which split from the main line in the first 100,000 years. An article in the Economist did a very good job of making the article understandable. Here is a quote from this terrific article in the Economist, which I suggest you read for yourself:
For more information, check out the following sources:
|
| Your personal health: The Personal Genome [business|bytes|genes|molecules] Posted: 24 Apr 2008 10:29 PM CDT
My take away from the discussion, which was fueled by questions submitted by the audience and via the web, was that there is so much uncertainty at this time. We know so much, yet so little. At some level, we do not understand the implications of what we know, ethical and medical, at the same time, we underestimate the ability of our own genetics to withstand changes. Perhaps one of the things that jumped out at me was the general popular belief (which is hardly surprising) that it is a gene or a few genes that can be altered or fixed to address a “problem”. We’re just beginning to grasp the relevance of pathways, of epigenetics, etc, so the long term implications of what we know (and don’t) are still a little fuzzy. I didn’t get a chance to record or take notes, but I was Twittering the whole thing. Unfortunately, I forgot to use a hashtag, which was silly. Much of the backchannel discussion was on the subject of designer babies. Leena Peltonen made some good points about the impact of genetic selection (we will not be able to alter germ lines), and whether it was desirable from the evolutionary perspective. George Church pointed out that people were going to do it anyway, much as they do today for sex selection or during IVF. Bill Gates had an interesting opinion on the question of what a personal genome really means. He differentiated between an individual with money getting themselves genotyped or sequenced and between the ability to sequence individuals cheaply and in large quantities. The latter for him was much more important since it will help advance science and medicine. He is quite right of course, and it will be interesting to see how the ability to sequence individuals cheaply has an impact on research and clinical studies, where, in theory, in a few years it will be possible to just sequence everyone. More on Sandra Porter’s blog Image via Wikipedia Technorati Tags: Personal Genomics, Personal Genetics, Ethics |
| Improve Your Financial Health, Reduce Stress and Help Save the Planet [Highlight HEALTH] Posted: 24 Apr 2008 09:30 PM CDT With crude oil hitting a record high this week, gas prices here in the U.S. are soaring. According to CNN.com, the $100 fill-up has arrived in the United States. Want to reduce your stress level, spend less money at the pump and do your part to help save the planet? Here’s one of the most simple yet effective tips that will accomplish all three: Slow Down. Improve Your Financial Health: Use Less Gas At highway speeds, wind resistance increases exponentially and fuel economy is reduced by approximately 4 miles per gallon for every 10 mile per hour increase [1]. Thus, the faster you drive, the more it will cost you. Consider this [1].
Additionally, if you do the math, speeding doesn’t save you anywhere near the time you might think it does. An average 30 mile commute traveling at 65 miles/hour takes 28 minutes, while that same trip at 80 miles/hour takes 23 minutes. You save a whole 5 minutes by driving 15 MPH faster. How much money is that 5 minutes worth? Reduce Stress and Stay Safe Moderate levels of stress from a variety of sources, including other motorists, traffic congestion and roadway conditions, are common in everyday driving. However, driver stress has been shown to also be influenced by a combination of situational and personal factors, including factors external to the driving context [2]. Not surprisingly, studies have found that life stress is associated with higher rates of accidents and disease [3]. It’s been estimated that drivers who have experienced a recent stressful event are five times more likely to cause fatal accidents than unstressed drivers [4]. If you’re running late, remember that no matter how fast you drive, you’re still going to be late. If you’re under a great deal of personal stress, it’s probably best to avoid driving altogether. Statistically, people who drive too fast cause or contribute to almost one-third of all fatal crashes. In 2006,13,543 lives were lost in speeding-related crashes [5]. Excessive speed does a number of things:
Want to reduce your stress level, spend less money on gas and do your part to help save the planet? Just Slow Down! David over at The Good Human has some additional tips on saving money, saving fuel and saving the environment. References
.gif) Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership! Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership!This article was published on Highlight HEALTH. Related articles |
| Timely Release and A Unanimous Vote [The Gene Sherpa: Personalized Medicine and You] Posted: 24 Apr 2008 08:49 PM CDT |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |









 Yesterday, a very interesting paper was published in the American Journal of Human Genetics by the Genographic Project Consortium entitled “
Yesterday, a very interesting paper was published in the American Journal of Human Genetics by the Genographic Project Consortium entitled “

 That’s based on a $3.25 price per gallon, which is less than the current price of gas. So we’re talking more than 54 cents a gallon.
That’s based on a $3.25 price per gallon, which is less than the current price of gas. So we’re talking more than 54 cents a gallon.
No comments:
Post a Comment