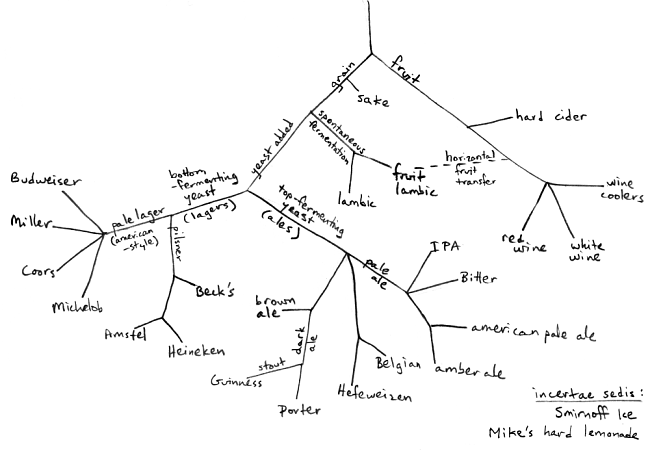
The DNA Network |
| Posted: 14 May 2008 08:37 PM CDT
via Tipping the Spherical Cow. .gif) Related posts | ||||||
| Canada Wide Science Fair [Bayblab] Posted: 14 May 2008 08:23 PM CDT For the past two days, I've been a judge at the Canada Wide Science Fair here in Ottawa. The students and their projects have been extremely impressive. For those in the Ottawa area who want to see some of the future stars of Canadian research and innovation, there is a public viewing this Saturday, May 17th between 9am and noon in Montpetit gym at the University of Ottawa. | ||||||
| Posted: 14 May 2008 06:14 PM CDT Just a little FYI. I was stumped today on what exactly an octane rating on gasoline meant. The octane rating is based on results using a test engine. It is the resistance to autoignition that is being measured. Autoingition is undesirable in a gasoline engine as this produces knocking. It is called an octane rating as iso-octane is defined as 100 and n-heptane is defined as 0. So a mixture of 87% iso-octane and 13% n-heptane would have an octane rating of 87%. However that doesn't mean that a fuel with a 97 octane rating has 97% octane, it just has characteristics as if it was 97% octane and 3% n-heptane. | ||||||
| Posted: 14 May 2008 05:32 PM CDT Thanks to Doug Gurian-Sherman, senior scientist with the Union of Concerned Scientists) for his thoughtful comments. My response here: 1. In our book, we make the point that sometimes GE is the most appropriate technology to solve a particular problem but not always. Geneticists, breeders and farmers are endlessly innovative and there are often many approaches to tackle a particular problem. There are cases where GE is the most appropriate technology. Case in point: GE papaya. There was no other tool available to combat papaya ringspot virus, nor is there now. Please see previous blog on this subject. 2. Doug says that it is not simple to find and commercialize a GE trait. I agree. All new varieties (GE or otherwise) need to be rigorously tested for their agronomic properties. This takes time and resources. This doesn't mean we should stop developing new varieties (GE or otherwise) though. 3. I agree that "the rising problem of herbicide resistant weeds and its consequences, for example, are a significant environmental (and practical) problem". But this is a problem anytime an herbicide is used extensively and is not an environmental consequence of GE. The same result would occur if the crop is GE or non-GE (and many such herbicide tolerant non-GE crops have been generated). GE crops themselves have had had no negative environmental or health effects after over 1 billion acres have been planted. 4. In some of its commercial forms, glyphosate is mixed with a compound called a surfactant that allows it to be more eff ective. Although glyphosate is nontoxic to freshwater fish, there is evidence that a surfactant called POEA, used in some formulations, is more toxic than glyphosate alone to aquatic species. If the surfactant or herbicide creates environmental problems then it makes more sense to simply regulate the surfactant or even the herbicide itself. 5. Bt has had some real benefits in reducing chemical insecticide use. The recent analysis by Cattaneo et al (2005) clearly shows that there is similar biodiversity (ants and beetles) in GE cotton fields vs non-GE cotton fields. In contrast, broad-spectrum insecticides (which are used on the vast majority of non-GE cotton fields throughout the world) significantly reduces ant and beetle species richness. 6. Although BT-cotton in China has been dramatically effective in reducing pesticide use, after 7 years some farmers started spraying again to control secondary insect pests that are not controlled by Bt. This points to the need to integrate GE crops into organic farming systems that also use crop rotation and beneficial insects to control secondary pests. Please see this article for more details. 7. "I don't see how you would fit herbicide tolerance into organic in any case". I agree. Herbicide tolerant crops were developed for conventional farming systems in the US and do not benefit organic farmers or most farmers in less developed countries that cannot afford the herbicide. Each new GE crop variety must be evaluated on a case by case basis. 8. As to negative health effects, while there may be none from current crops, we really don't have the data to know with much certainty. Please see article by Engber. 9. "The FDA process is voluntary, with no required tests and very few recommended ones". As far as I know all US companies that have released transgenic crops have submitted their new variety to the FDA process. 10. "The claims by many of your more vocal colleagues that these crops are more stringently regulated than any others is moot because the standard of comparison for many of these crops is nothing! I.e., other crops are not regulated at all for safety, so of course ANY regulation, even the weakest, is by comparison more stringent!" I agree that GE crops are regulated whereas other crops are not. Doesn't this kind of regulation indicate much greater stringency? 11. " The question should not be whether a few current transgenic traits out of dozens or hundreds are harmful, if the technology goes forward as many would like, but whether the regulations are adequate to reliably detect those that may be harmful". I agree. Although there is wide consensus here in the US and abroad that all GE crops currently on the market are safe to eat, all future crops must be evaluated on a case by case basis. 12. "Although you have alluded to some of the big issues around GE in your other posts, you don't deal with them head on (again, apologies if you do that in you book)". Please see chapters 10 "who owns the seed?" and chapter 11 "Who owns the genes?" in Tomorrow's Table. 13. "Finally, you do not address the very important issue of opportunity costs when pursuing GE. In a world of limited resources, we need to put those resources where they can do the most good." Current interventions to combat vitamin A deficiency rely mainly on pharmaceutical supplementation, which is costly in the long run and only partially successful. The most recent estimates indicate that golden rice is expected to save lives of thousands of children. This is a golden opportunity to use GE. Lives of children are at stake. | ||||||
| Posted: 14 May 2008 05:15 PM CDT It is coming. Yes, 23andme is challenging the traditional way we are conducting clinical trials. In a press release today, 23andme is announcing a partnership with the the Parkinson's Institute to discover genetic and environmental factors of Parkinson's disease. Six months ago, my colleagues and I send out a grant proposal arguing the potential efficiencies of combining consumer genomics with clinical trials. Although I am frustrated to learn a week ago that our proposal was not funded, I am very happy to see the press release today from 23andme, which essentially validated our proposal.
Similar to the early days of sequencing and bioinformatics development, I would expect to see the industry driving the innovative applications, instead of the academics. There are "2.0" hypes on everything recently, including the "Research 2.0" in the press release. Even though there are a lot of details to be worked out, I am still very positive on it. | ||||||
| Dr Watson's Genetic Counselor: Witty or Insulting? [PredictER Blog] Posted: 14 May 2008 04:42 PM CDT Today's issue of Nature [subscription required] includes a letter responding to Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872-876 (17 April 2008) | doi:10.1038/nature06884. The author of the response, a genetic counselor, lifts a layer off the science publishing hype that surrounds anything about the human genome in this era. Also see the very insightful and witty table comparing two potential personal genome and genetic counseling clients: "Dr. Watson" and a "lay patient". Here's a sample: - Dr. Watson: Thinks the $1 million cost is a good deal - Lay Patient: Worried about the cost of a - consultation - Dr. Watson: Brings in sequence data on a hard drive - Lay Patient: Brings in records about sinus infections - Dr. Watson: Chose to have Apo-E sequence redacted - Lay Patient: Expects to learn blood type - Dr. Watson: Shares 1.68 million SNPs with Craig Venter - Lay Patient: Googles SNPs to find out who they are Well said! But gee, the "lay patient" must be a real dimwit ... if I ever need a genetic counselor, I'm going to do my homework first! - J.O. Source: Roche MI. A case of genetic counselling for Dr Watson. Nature 453, 281 (15 May 2008) | doi:10.1038/453281a; Published online 14 May 2008. | ||||||
| How is a starving, mutant yeast like a cancer cell? [adaptivecomplexity's column] Posted: 14 May 2008 03:11 PM CDT When yeast are starving, they do what any rational microbe would do: they stop dividing and hunker down, trying to conserve resources until the good times return. | ||||||
| Posted: 14 May 2008 02:00 PM CDT I can officially claim myself as a member of the ivory tower elite. At least, that's what they tell me. Read the comments on this post... | ||||||
| X-Prizes for Health and Medicine? [ScienceRoll] Posted: 14 May 2008 12:21 PM CDT Have you ever heard about the Ansari X Prize that resulted in constructing the world’s first privately developed spacecraft? Have you ever heard about the Archon X-Prize for Genomics? It will lead us to a new generation of genome sequencing methods. What about an X Prize for Health and Medicine? Students at the Massachusetts Institute of Technology came up with some interesting ideas.
What do you think? An X Prize for Health and Medicine would be a good idea? Which medical specialty would be your choice?  | ||||||
| Sequence and Ligation Independent Cloning [Bitesize Bio] Posted: 14 May 2008 07:31 AM CDT Regular readers will know about the advantages of T4 DNA polymerase-mediated ligation independent cloning. The fact that it is faster, more efficient and allows easier parallel cloning than conventional cloning has made it my method of choice in the lab. But the technique does have it’s downsides - not least the requirement that existing vector multiple cloning sites be modified to convert them into ligation independent cloning vectors. This paper by Li and Elledge recently flagged up in a comment by Max (thanks Max!) looks like it could change all that. It turns out that no sequence modification is required at all and LIC (or sLIC - sequence and ligation independent cloning as the authors call it) can be performed at any site in any vector of your choice. If you are familiar with T4-mediated ligation independent cloning you will know that the vector sequence needs a specific LIC site containing restriction site flanked by regions that lack one of the nucleotides (e.g. adenine) for a 13-14nt stretch (read this first if you are not familiar with it). After linearising at the restriction site, the vector is incubated with dATP + T4 DNA polymerase, which chews back the 3′ end of the DNA until it stops after 13-14 nucleotides due to the presence of the dATP. This creates a single-stranded region to with a similarly treated insert can be annealed. In this paper, Li and Elledge showed that the specific vector LIC site was not required. Treating the vector with T4 DNA polymerase and no dNTPs for a certain length of time (30 minutes was optimal for them) was sufficient for vector preparation. The single stranded stretch this creates is longer than required for annealing an insert, but single stranded gaps like these are apparently repaired very efficiently by E.coli after transformation so this is not a problem. They also demonstrated that inserts prepared in one of three ways could be successfully annealed and transformed, which considerably increases the versatility of the process. The insert prep methods were: 1. T4 DNA polymerase treatment. Just like the vector, the insert could be subjected to T4 DNA polymerase treatment (without dNTPs) to create single stranded regions that will anneal to the prepared insert. The authors showed that efficient annealing needed only 20-30 nt (single stranded) regions of homology at each end of the vector and insert. Amazingly, the homologous regions didn’t even need to be at the ends of the insert - the authors showed that non-homologous regions of up to 20 nucleotides could be tolerated as the branched products produced after annealing are efficiently trimmed and repaired by the cell. I have not had a chance to try out this method yet, but it certainly looks very exciting. It looks to me like once this I have this protocol is up and running there will be no need at all for restriction enzyme-mediated cloning… which is a day I long for! When I have some results of my own from this I’ll publish them, and my protocol here so watch this space. | ||||||
| Genetic Manipulation [Sciencebase Science Blog] Posted: 14 May 2008 07:00 AM CDT
Are you happy to eat genetically modified foods? What about your friends and colleagues? Do the GM pros outweigh the cons? I asked a few contacts for some answers by way of building up to a more formal response to those kinds of questions that will be published soon in the International Journal of Biotechnology (IJBT, 2008, 10, 240-259). Plant geneticist Dennis Lee, Director of Research at mAbGen, in Houston, Texas, suggests that GM crops have several significant advantages. “Total cost per acre can actually be significantly less for GM crops,” he says. This is particularly true for crop species, such as maize, that have been modified to produce natural toxins that fend off insect pests or protect the crop from the herbicides need to keep weed growth at a minimum. However, he points out that, “In practice, this is often not the case - farmers tend to err on the side of caution and continue to use significant amounts of pesticides and herbicides.” That said, crops can also be modified to grow in substandard conditions, such as strains of tubers grown in Kenya that are capable of surviving both drought conditions and high-salt soils. “Obviously, this is beneficial to yield - you can actually get some food out of places where you previously could not,” adds Lee. In addition, it could be possible to modify some crops to have greater nutritional content, such as the so-called “golden rice” project by Ingo Potrykus then at the Institute of Plant Sciences of the ETH Zurich. One of the biggest perceived problems regarding GM crops is the possible contamination of other species. What if Jeff Chatterton, a Risk and Crisis Communications Consultant at Checkmate Public Affairs, in Ottawa, points out that the pros are well documented: increased yield per acre, ease of use and perhaps, some day, increased ‘consumer level’ benefits such as higher nutritional values. But, echoes others’ comments on the hidden con of farmers the world over potentially being locked into the agbiotech company’s seed and having no recourse to produce their own from one year to the next. “As traditional family farms are increasingly moving towards “Roundup Ready” corn or soybeans, you’re increasingly seeing a change in the business model of farming,” he says. “Rather than ‘family farms’ using traditional farming practices, agricultural operations are increasingly becoming factory farms.” It might be said that the emergence of factory farms is occurring outside the realm of GM crops, but with pressure being applied to produce more and more crops for non-food purposes, including, biofuels, unique polymers, and other products, the notion of a factory farm that doesn’t even feed us could become an increasing reality. Lee also mentions an intriguing irony regarding the public perception of risk-benefits concerning GM crops and that is that the toxins produced by modified Bt maize is exactly the same toxin produced by the natural soil microbe Bacillus thuringiensis (Bt) itself and this is same Bt toxin that so-called “organic” farmers are usually allowed to use instead of “synthetic” pesticides. Information Technology and Services Professional Bill Nigh of Bluenog, based in New York, provides perspective as a lay person. “We’ve been engaged in genetic manipulation for a long time now,” he says, “but it was limited by the technology at hand. With recombinant DNA it’s a remarkably more vast All that said, an international team has now investigated the various issues and has assessed the public’s Willingness to Accept (WTA) GM foods based on experimental auctions carried out in France, UK, and USA. Lead author of the IJBT paper Wallace Yee now at the University of Liverpool, worked, while at Reading University, with colleagues in various disciplines, from agricultural and food to business and economics in Italy, New Zealand, UK and US to explore perceptions of risk and benefits, moral concerns and attitudes to the environment and technology. “Trust in information provided by industry proved to be the most important determinant of risk/benefit perceptions,” the researchers conclude, “willingness to accept followed general attitudes to the environment and technology.” They also found that educational level and age could also enhance perceived benefits and lower perceived risks of GM foods. “Our research suggests that trust-building by industry would be the most effective approach to enhancing the acceptance of GM foodstrust-building by industry would be the most effective approach to enhancing the acceptance of GM foods,” the team says. “If the industry could educate people that GM technology does not pose any threat to the environment, but provides benefits to society as a whole and consumers as individuals, the attitudes of the public towards GM in food production would be favourable, and in turn increase their willingness to accept,” they conclude. Computing professional Paul Boddie of Oslo, Norway, coming at the issue of GM crops from an indirect angle provides an allusion to computer programming that seems quite pertinent and was originally attributed to Brian Kernighan, which Boddie suggests readily transfers to other disciplines including genetic engineering: “Everyone knows that debugging is twice as hard as writing a program in the first place. So if you are as clever as you can be when you write it, how will you ever debug it?” A post from David Bradley Science Writer | ||||||
| What does DNA mean to you? #5 [Eye on DNA] Posted: 14 May 2008 03:03 AM CDT Reader jhay of The Four-eyed Journal says:
| ||||||
| TED talks: Space [Mailund on the Internet] Posted: 14 May 2008 02:00 AM CDT I’m a bit of a space exploration enthusiast myself, so of course I have to list a few TED talks on that as well… The future of space flightExploring Earth and the solar systemI would love to see this happen… This might be my favourite talk of them all. I was almost applauding myself at the end. Project OrionThis was a really cool project that never happened (for good or bad… atomic rockets are a bit risky but very powerful). Our place in the cosmos…and really a lot more than cosmology… World wide telescopeI wasn’t sure if this should be under the heading “Space” or “IT”, but here it is… Table of contents for TED Talks
| ||||||
| Ubiquitous computing [business|bytes|genes|molecules] Posted: 13 May 2008 08:10 PM CDT
I want to cast that problem a little differently. Let us assume we have a scalable cloud that we can tap into, perhaps with databases that easily scale to support streaming data. Let us imagine a lab, with instruments streaming out data, with devices consuming those data, and systems that can make decisions based on the data they are receiving. Such an interconnected work, a world with pervasive, ubiquitous computing might sound like science fiction, but over the past few years, we have slowly but surely built up the beginnings of an infrastructure that will make this scenario possible, perhaps a lot faster than we thought. The iPhone, the Chumby, the Bug, these are just early examples, as are streaming video services and communication platforms like Twitter and XMPP. Of course, before we get there, we really need to develop systems that are smart about making decisions and filtering information, otherwise, we’re just going to get buried in a deluge of data that will make our heads spin, individually and collectively. Update: On a semi-related note, here’s an Ignite talk from Where 2.0 that is an example of where we are headed (via O’Reilly Radar)
Further reading Image via Wikipedia Technorati Tags: Ubiquitous Information, Streaming Data | ||||||
| Book Review: Beyond Genetics by Glenn McGee [Think Gene] Posted: 13 May 2008 08:07 PM CDT Beyond Genetics is a core of a few short essays fluffed into a book. Everything about this book screams “move my consulting career” including:
Some of the book I liked. For instance, McGee does a good job of discussing the gene patent debate. But this book was clearly written to sell in popular bookstores and boost McGee’s stature in a fledgling industry. As such, Glenn is controversial only enough to be noticed, and he strokes every political group in the genomic industry without assuming any thoughtful responsibility for his genomic vision because “hey, it’s the future, right?” So if he’s wrong, this book will be safely forgotten without offending anyone, and that’s what’s already happening. If you liked “The World is Flat,” you may like this book. Otherwise, let this one collect dust at the Barnes and Nobles. | ||||||
| Scientific Research and Medicine in Second Life [ScienceRoll] Posted: 13 May 2008 04:35 PM CDT David Pescovitz at BoingBoing had an interesting post about scientific research in Second Life, the virtual world:
This is a great example about how to close the gap between real science and the virtual world. You may also know about our medical exercises at the Ann Myers Medical Center where they train medical students (just like me) with case presentations. Now here is a tool with serious potential that could help us do it even better: Text files, images and videos in one place. The aim is to create virtual patients with virtual medical conditions. Why is it beneficial for medical students? The entire medical education is about case presentations. We move forward clinic by clinic during our studies and listen to case presentations all the time. That’s what we can do in Second Life but with even more educational material and without time or geographical restrictions. According to DusanWriter, there are pharmacy patient case studies as well in Second Life.
If you want to know everything about Second Life & Virtual Worlds for Academic Healthcare & Education, check out the slideshow of Patricia F. Anderson. And if you don’t have a computer at home, let’s use a mobile to enter Second Life: Further reading:
 | ||||||
| DDC Helps Unite Family Through DNA Testing for Immigration [The DNA Testing Blog] Posted: 13 May 2008 03:59 PM CDT This year, DDC helped make Mother's Day very special for one Ohio family. The Ezenagus are originally from Nigeria but now live together in a Northern suburb of Cincinnati, and it has taken over a decade for all members of the family to secure their visas, or "green cards," and immigrate to the United States. For [...] | ||||||
| X2, a hypothesis aggregator, is surprisingly interesting and engaging. [Synthesis] Posted: 13 May 2008 03:34 PM CDT My colleague Attila pointed me to X2, an effort by the Institute for the Future to collect and collaboratively rank hypotheses about future directions of science. When I read about it, it sounded interesting, but coming from futurists, I rather expected it to be all style and no substance. I was pleasantly surprised to find a substantial amount of interesting content on the site, and it seems like the core functions all work as expected. Instead of being just another pawn in my attempt to own the first page of Google results for my name, William Gunn, (I dominate Mr. Gunn already The basic function of the site is similar to Scintilla, in that users submit content and you can rate it and find related content you may not have seen, but instead of being fed by a collection of RSS feeds .gif) This post is about future, hypothesis, prediction, recommendation, science This post is about future, hypothesis, prediction, recommendation, scienceRelated posts | ||||||
| cyclone Nargis: do we let them stand alone? [the skeptical alchemist] Posted: 13 May 2008 03:06 PM CDT  If you are a fan of the Lord of the Rings, you might just remember that moment when you see the Elves arriving at Helm's Deep, at a time when the Rohirrim thought that nobody would ever come to help in the fight against Saruman. I remember well, because so many things happen in this world when that moment comes again and again, and nobody ever decides not to let people in need stand alone. Rwanda and Darfur come to mind. Man-made wars. If you are a fan of the Lord of the Rings, you might just remember that moment when you see the Elves arriving at Helm's Deep, at a time when the Rohirrim thought that nobody would ever come to help in the fight against Saruman. I remember well, because so many things happen in this world when that moment comes again and again, and nobody ever decides not to let people in need stand alone. Rwanda and Darfur come to mind. Man-made wars.But now, here comes this case. This case is, in most people's minds, so unequivocal, that our inability to react to it will be perceived by most as deeply unethical. Here comes a time when, struck by the force of nature, a government fails an entire nation...and we stand here to watch and pontificate on whether it would be legal for the UN to send troops to Myanmar. Too bad we know exactly why Russia and China voted against a UN resolution in favour of stronger UN intervention in Myanmar (two words: Tibet and Chechnya).
Apparently, it can also take about 3 hours for a European national on Burmese ground to get a tourist visa, but if you happen to be a relief worker, you will simply not be allowed in. I guess this is what the ruling junta wants, ahead of the upcoming constitutional referendum which could keep them in power for a very long time still. Which makes me wonder whether the area struck by the cyclone would have voted, before the events, against or in favor of the junta remaining in power. Never underestimate a bureaucracy's instinct for self-preservation. View blog reactions | ||||||
| Research is a money pit [Bayblab] Posted: 13 May 2008 12:11 PM CDT Do you ever get the sense that research money often goes wasted? Even if you ignore all the research dead-ends and the discoveries which have limited implications, and just concentrate on what you can get for your tax money. How often does an expensive machine get used for only one experiment, how often do we throw out stuff that still works. And not to mention the price gouging that suppliers are guilty of. I don't understand how a ice pack for a western apparatus can be 10 times more expensive than a regular one, or why a research fridge is four times the price of a regular fridge. It's certainly not for the reliability, from what I've seen of our fridges. Some of the kits you can get now are ridiculous, especially considering the hourly wage of the person using it. It begs the question: should we hold the researchers accountable for the money they use? Between two researchers who produce the same work, should we choose the one who will do it for cheaper, or do we risk stifling innovation? Just take this example of a researcher who used his grant money for chrome wheels and big screen TVs. It sounds like he got off easy: "Another $123,703.20 in expenditures appeared to be "inconsistent" with his research grant proposals, the documents say. But university investigators gave him the benefit of the doubt saying the nine computer monitors and other items "might have been related" to his research from a "general scientific perspective." The university then made an arrangement with the researcher that in the "event of timely repayment" of $24,767.33, it would not ask for the rest of the money back. The university returned $21,485.67 to NSERC. The documents indicate the other $3,000 refunded by the scientist was sent back to other agencies that had also financed his research." | ||||||
| Everything You Wanted to Know About the Human Genome But Were Too Lazy to Figure Out [Bayblab] Posted: 13 May 2008 11:37 AM CDT This new book sounds cool. "A Short (173pp) Guide to the Human Genome", by Stewart Scherer. Probably a great one for your bathroom or outhouse collection. Such a reference is drastically needed - everyone in biology should know the human genome inside out, but who has the time to fish out all the info? Now we don't have to. Sweet. | ||||||
| The Epi-Genomic Canary [The Gene Sherpa: Personalized Medicine and You] Posted: 13 May 2008 08:51 AM CDT | ||||||
| Post-Modernism versus Science [Bitesize Bio] Posted: 13 May 2008 05:44 AM CDT
I came across an outstanding essay on the subject by Daniel Dennett over on Butterflies and Wheels: Postmodernism and Truth. There are a few excerpts below the fold [Emphasis mine]. Yes, there’s still Kuhnsian sociology of science, although I’ve always read Kuhns as agreeing with the bit from the last quote that I have below the fold: “The methods of science aren’t foolproof, but they are indefinitely perfectible. Just as important: there is a tradition of criticism that enforces improvement whenever and wherever flaws are discovered.”
Granted, scientists are still human and falliable. But yes, the whole of the endeavor of scientific research, to say nothing about other technical occupations, are centered around scrutinization of results, and “bootstrapping our way to ever greater accuracy and objectivity,” to refer to the next quote. I would just change accuracy to precision there, because accuracy implies a pre-defined target, whereas precision instead implies the reproducibility of results. As I found out early on in grad school, trying to fit your results to your expectations is bass-ackwards.
In case you haven’t noticed from reading scientific papers, researchers, amongst themselves, are notoriously conservative (not in the political sense) in how they make inferences based on available data. If you claim something in a manuscript, the peer-review process is supposed to call you out on it - and if the initial reviewers miss such overstatements, someone somewhere will probably notice, sooner or later. Funding requests are another thing, of course, but even there a scientist is judged on their ability to collect and interpret data which matches reality. If they fail to meet expectations in terms of data-driven research, then they’re in trouble.
Platonism may have been greatly flawed in some respects, but in mathematics, he and his fellow Ancient Greeks were very perceptive fellows. They noticed that quantification was as objective a way of interpreting reality as you could get. And such quantification is excactly what the precision of science is built on - exemplified best by the calculation of the value of Pi. We still don’t have an accurate or perfect number that equals Pi, only an astoundingly precise estimate for that number. The same is true for all of science - again, we are bootstrapping our way to better and better approximations of reality.
Indeed. And this epistemology is how knowledge is acquired (as opposed to certainty*). * = Science has proof without any certainty. Faith has certainty without any proof. | ||||||
| TED talks: IT [Mailund on the Internet] Posted: 13 May 2008 04:00 AM CDT Continuing on listing my favourite TED talks, I now turn to IT. Information technology is, in my view, the major innovation of the last decades. It is the enabler for all the exiting science we do these days, and the enabler for plenty of changes to our everyday life as well. Open SourceThe InternetWell, it isn’t so much the web, the last one, but a history of electricity and appliances … but it is remotely related to the .com boom. An optimistic view on it, even… Table of contents for TED Talks
| ||||||
| Posted: 13 May 2008 02:59 AM CDT Well, much as I hate to admit it, I find myself agreeing with the notion that cuteness sells in genome sequencing. That is in essence the claim of Natalie Anger in an article in the New York Times about all the attention the platypus genome paper has been receiving over the last week (see A Gene Map for the Cute Side of the Family - New York Times Alas, microbiologists really do not have anything like this no? I mean, who feels that E. coli or yeast are, well, cute? (Well, even if you have one of those "giant microbes" stuffed animals, that just means you are a dork like me ... the public does not collect those). Sure, Carl Zimmer can get some attention for all the geeky tattoos out there and some of them did have something to do with microbes, but again, a platypus they are not. So what are we forlorn microbiologists to do? We need better PR and imagery. We need cute microbes. We need more dark and evil microbes too (I mean, if anyone sequenced the T-rex genome - for real - it would get attention too). So - I am calling all microbiologists and microbiology fans --- bring forth your imagery that will help microbes get the attention they deserve. And today I am suggesting just one simple thing we can all do to make a difference: get some new names. That is, give your favorite microbe a good common name or nickname to bring out the cuddly or dark imagery we need. All microbes names should conjure up something to the public, like anthrax does (yes, I know, anthrax is the disease and not the microbe , but this adherence to rules is part of the problem we have). Here are some proposed name changes for organisms I have worked on: Wolbachia - "The Feminizer" Tetrahymena - "The Hairy Beast" Carboxydothermus hydrogenoformans - "Exploding Breath of Death" Chlorobium tepidum - "Little Green Machine" So - please - come up with nicknames for all your bugs and start to use them or at least post them here. | ||||||
| Think Gene Publishing Policy [Think Gene] Posted: 13 May 2008 01:45 AM CDT Hi readers and DNA Network. Thank you for your continued readership and community. Here at Think Gene, we’ve been busy writing new software and raising funds for a bio-related software startup. To keep the site populated with content, we’ve written a few tools to help us find, format, and publish what we think are the most interesting bio and genomic news items of the day. However, our tools were just a bit TOO good, and we’ve been publishing a bit more news than what is reasonable as a member of the DNA network. To address this issue, here is our new publishing policy:
Thank you for your patience while we update our tools and software to meet the new Think Gene publishing policy. We hope to have a few more exciting Think Gene announcements in the near future! As always, please do not hesitate to leave us feedback in the comments or by our email. -Andrew Yates | ||||||
| When you care enough to send the very best DNA [Omics! Omics!] Posted: 12 May 2008 10:32 PM CDT Yesterday was Mother's Day, and while searching for a card I spied what looked like a double helix on the front of one card. Finding this odd, I checked the card in detail -- and indeed it was DNA! DNA is clearly in the public consciousness -- years of Law & Order and CSI have ensured that, but I found it striking that the image of a double helix is deemed recognizable by as mainstream & middlebrow a company as Hallmark. A nice twist is the card actually bore a message along the lines of 'even though you didn't give me any DNA...' -- a card for mother figures, not birth mothers. So this isn't a sign of rampant DNA deterministic thinking, but rather the imprint of DNA on the public (or at least corporate) mind | ||||||
| Scientists dig deeper into the genetics of schizophrenia by evaluating microRNAs [Think Gene] Posted: 12 May 2008 10:29 PM CDT
In the May 11 issue of Nature Genetics, Maria Karayiorgou, M.D., professor of psychiatry, and Joseph A. Gogos, M.D., Ph.D., associate professor of physiology and neuroscience at Columbia University Medical Center explain how they uncovered a previously unknown alteration in the production of microRNAs of a mouse modeled to have the same chromosome 22q11.2 deletions previously identified in humans with schizophrenia. "We've known for some time that individuals with 22q11.2 microdeletions are at high risk of developing schizophrenia," said Karayiorgou, who was instrumental in identifying deletions of 22q11.2 as a primary risk factor for schizophrenia in humans several years earlier. "By digging further into this chromosome, we have been able to see at the gene expression level that abnormalities in microRNAs can be linked to the behavioral and cognitive deficits associated with the disease." The investigators modeled mice to have the same genetic deletion as the one observed in some individuals with schizophrenia and examined what happens in the expression of over 30,000 genes in specific areas of the brain. When they discovered that the gene family of microRNAs was affected, they suspected that the Dgcr8 gene was responsible. The Dgcr8 gene is one of the 27 included in the 22q11.2 microdeletion and has a critical role in microRNA production, so this was a logical hypothesis. Indeed, when they produced a mouse deficient for the Dgcr8 gene, and tested it on a variety of cognitive, behavioral and neuroanatomical tests, they observed the same deficits often observed in people with schizophrenia. The significance of this work is that it implicates a completely novel, previously unsuspected group of susceptibility genes and brings investigators a step closer to understanding the biological mechanisms of this disorder. Implication of such a large family of genes (the most recent estimate puts the number of human microRNAs at at least 400 that influence the expression of as many as a third of all genes) could partly account for the genetic complexity associated with this devastating disorder and explain some of the difficulties that the researchers have encountered in their efforts to pinpoint individual genes. "Our hope is that the more we know about the genes involved in schizophrenia, the more targeted treatment can be," said Dr. Gogos. "Much in the way that cancer patients who have tested for a particular gene, such as BRAC1, can be tested and then treated with protocols designed specifically for them, we want to be able to know enough about the schizophrenic brain to target treatments to individual patients." The next step for the researchers is to find the many genes whose expression is controlled by the identified deficient microRNAs, which could in turn be involved in the pathogenesis of schizophrenia. Much more study and identification of other genetic variants must be done to further illuminate the disease's genetic underpinnings, according to Drs. Karayiorgou and Gogos. Source: Columbia University Medical Center | ||||||
| Human aging gene found in flies [Think Gene] Posted: 12 May 2008 10:28 PM CDT Scientists funded by the Biotechnology and Biological Sciences Research Council (BBSRC) have found a fast and effective way to investigate important aspects of human ageing. Working at the University of Oxford and The Open University, Dr Lynne Cox and Dr Robert Saunders have discovered a gene in fruit flies that means flies can now be used to study the effects ageing has on DNA. In new work published today in the journal Aging Cell, the researchers demonstrate the value of this model in helping us to understand the ageing process. This exciting study demonstrates that fruit flies can be used to study critical aspects of human ageing at cellular, genetic and biochemical levels. Dr Lynne Cox from the University of Oxford said: "We study a premature human ageing disease called Werner syndrome to help us understand normal ageing. The key to this disease is that changes in a single gene (called WRN) mean that patients age very quickly. Scientists have made great progress in working out what this gene does in the test tube, but until now we haven't been able to investigate the gene to look at its effect on development and the whole body. By working on this gene in fruit flies, we can model human ageing in a powerful experimental system." Dr Robert Saunders from The Open University added: "This work shows for the first time that we can use the short-lived fruit fly to investigate the function of an important human ageing gene. We have opened up the exciting possibility of using this model system to analyse the way that such genes work in a whole organism, not just in single cells." Dr Saunders, Dr Cox and colleagues have identified the fruit fly equivalent of the key human ageing gene known as WRN. They find that flies with damage to this gene share important features with people suffering from the rapid ageing condition Werner syndrome, who also have damage to the WRN gene. In particular, the DNA, or genetic blueprint, is unstable in the flies that have the damaged version of the gene and the chromosomes are often altered. The researchers show that the fly's DNA becomes rearranged, with genes being swapped between chromosomes. In patients with Werner syndrome, this genome instability leads to cancer. Cells derived from Werner syndrome patients are extremely sensitive to a drug often used to treat cancers: the researchers show that the flies that have the damaged gene are killed by even very low doses of the drug. Professor Nigel Brown, Director of Science and Technology, Biotechnology and Biological Sciences Research Council said: "The ageing population presents a major research challenge to the UK and we need effort to understand normal ageing and the characteristics that accompany it." "Fruit flies are already used as a model for the genetics behind mechanisms that underlie normal functioning of the human body and it is great news that this powerful research tool can be applied to such an important area of study into human health." Source: Biotechnology and Biological Sciences Research Council | ||||||
| Model shows how mutation tips biochemistry to cause Alzheimer’s [Think Gene] Posted: 12 May 2008 10:28 PM CDT Your fate can be determined by tiny events. Imagine you live in the city and you walk everywhere to get exercise – you are healthy and not afraid of getting mugged. You almost never eat breakfast so you don't stop at the donut shop on the way to work, until one day the manager replaces the girl at the counter with her pretty red-haired younger sister. This seemingly unimportant change in your world is just enough to overcome your ability to resist high-fat temptation. A million donuts later, your cholesterol level surges and then your heart gives out. Curse you, little red-haired girl! Like staff change at the donut shop, subtle, seemingly inconsequential differences in human genetic design can lead to some unexpected tipping points in cellular chemistry that can lead to disaster. Cellular processes, like all the routines of life, are unfathomably complex, constantly evolving, and are sometimes dramatically sensitive to the smallest of changes. Consider the case of Alzheimer's disease… Alzheimer's is a terrifying brain-destroying disease whose causes have proven very difficult to pin down. In recent years, science has been closing in on solving the puzzle, particularly regarding some of the hereditary, "early onset" forms of the illness. Unusual by-products of cell metabolism, clumps of protein aggregates, have been shown to have a toxic effect on brain cells and certain gene mutations have been shown to be associated with increasing production of these by-products, though the evidence for an exact mechanism has remained hidden. Now, using sophisticated computer simulations, a team of physical chemists have shown precisely how a minor, seemingly inconsequential mutation results in unexpected changes in a very delicate chemical balance, creating build-up of the toxic by-products. The mutation, the substitution of a single base among the 3 billion found in human DNA, seems to have the greatest effect on a fragment of a specific protein that is abundantly present in living cells. The difference causes a subtle change in the shape of the fragment at a critical point, which can slightly shift the odds towards an inappropriate biochemical reaction that sidetracks the metabolic path. The increase in the reaction simply tips the balance of chemical processes, causing the build-up of a substance that kills brain cells, leading to the early deterioration of mental capacity and, eventually, death. "It is a really tiny change but it has tremendous consequences," said Andrij Baumketner, lead author on the study and a faculty member in the department of physics and optical science at the University of North Carolina at Charlotte. The finding, published in the April 7 issue of the Publication of the National Academy of Sciences, was co-authored by Mary Griffin Krone and Joan-Emma Shea, both from the department of chemistry and biochemistry at the University of California at Santa Barbara. The group studied the effects caused by the Dutch Mutation, a mutation that has been discovered to be associated with a specific, hereditary form of Alzheimer's disease. The mutation is small, the simple substitution of one DNA base for another, resulting in the change of only one amino acid residue – glutamic acid changing to the very similar glutamine – among hundreds of amino acids that form a protein known as the amyloid precursor protein (APP). The greatest effect of the Dutch-type mutation on APP, whose primary biological function is unknown, seems to be through a fragment known as amyloid-beta peptide that is created when cells break down the protein. Studies have shown that mutated forms of the fragment have greater tendency to stick to bond together and form protein clumps or aggregates. Some forms of the amyloid-beta clumps have been shown to be toxic to brain cells. Why the change in one amino acid would cause this peptide to form clumps more readily has, until now, been unclear. Amyloid-beta peptide, unlike most other proteins present in the cell, is largely lacking in specific shape (conformation), the characteristic that usually controls how proteins interact with each other. However the fragment does have two places in its sequence of amino acids – a section known as the "bend" and an area known as the "central hydrophobic cluster" where the polypeptide chain does conform to a more-or-less fixed shape. These areas, in fact, seem to be involved when the fragments bond together into clumps. The researchers created complex computer models of the two structured areas of the fragment and found that the single amino acid change caused by the mutation had a subtle effect on their properties. In order for fragments to bond together, the structured areas must first undergo a "conformational change" (a change in structural shape) from the conformations they normally have as single, water-soluble amyloid-beta peptides into a "transition state" conformation that leads up to forming clumps. The researchers found that mutation increased the likelihood that the structures would be in a form similar to the transition state before the reaction occurred. When the structured areas were already in the required transition state, bonding was encouraged because less energy was required for the bonding reaction to take place. "We knew quite a bit about what these peptides are from experimental studies, but we didn't know the microscopic details," noted Baumketner. "An experiment never gives you atomic resolution – you always have to guess what is actually going on with the molecules. But with a computer simulation you start with atoms and how they interact and you end with atoms, so there is no question with missing any details." The detail of the simulations showed that, because the mutation made the protein fragments more likely to be in a transition state for bonding, bonds between fragments were more likely to be formed than broken (in the reverse reaction), so clumps of fragments accumulated. The end result of the subtle, mutation-driven change in the protein fragment's shape was the tipping the reaction's balance enough to allow clumps composed of multiple fragments to occur and to build up –with a disastrous effect on brain tissue. "The barrier between the reactants and the products is the conformational difference between the peptide and its transition state," Baumketner said. "The fewer changes the peptide needs to undergo, the easier it is for it to change. The mutation predisposes the single amyloid-beta peptide to jump onto the barrier that keeps the reaction from happening." The ultimate problem responsible for Alzheimer's Disease, Baumketner notes, is that the design of the protein affected is so "close to the edge" in the reactions it must undergo that extremely small changes can cause problems, like the formation of toxic by-products. "It looks like whoever designed the proteins in our bodies only made the beta peptides to be right on the edge of where they have to be for us to be alive," he said facetiously. "You make a small push and you push it over the edge and then there is no return. If you were farther from the edge, that would be fine, and you could tolerate one mutation. "There is lots of discussion about why this happens – is it the failure of evolution? Maybe evolution never has had a chance to optimize us against this. Humans now live to be much older, but evolution never has had a chance before to detect and avoid these problems through natural selection. When the lifespan was 35 years, you didn't have a large problem with Alzheimer's. Now you do." Source: University of North Carolina at Charlotte |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |










No comments:
Post a Comment