The DNA Network |
| Posted: 20 Jun 2008 07:40 PM CDT James Schummers, Hongbo Yu and Mriganka Sur present their measurements of Ca++ dynamics in response to visual stimuli in the awake ferret and reveal highly refined patterns of astrocyte activity in their paper, “Tuned Responses of Astrocytes and Their Influence on Hemodynamic Signals in the Visual Cortex” (DOI: 10.1126/science.1156120). The genetic regulation of neurovascular coupling is key to understanding the way in which genetic variation may regulate brain function (or at least function as measured by the BOLD response). A closer look at BOLD response and genetic pathways that mediate astrocyte function would be music to my ears - or at least my auditory cortex astocytes. |
| Guest Post Today, Hopefully he doesn't get Flamed! [The Gene Sherpa: Personalized Medicine and You] Posted: 20 Jun 2008 05:48 PM CDT |
| Posted: 20 Jun 2008 04:19 PM CDT A small molecule that locks an essential enzyme in an inactive form could one day form the basis of a new class of unbeatable, species-specific antibiotics, according to researchers at Fox Chase Cancer Center. Their findings, highlighted on the cover of the June 23 issue of the journal Chemistry & Biology, take advantage of an emerging body of science regarding “morpheeins” – proteins made from individual components that are capable of spontaneously reconfiguring themselves into different shapes within living cells. The researchers discovered a small molecule, which they have named morphlock-1, binds the inactive form of a protein known as porphobilinogen synthase (PBGS), an enzyme used by nearly all forms of cellular life. The functioning form of PBGS is built from eight identical component parts – in what is called an octamer configuration – and is essential among nearly all forms of life in the processes that enable cells to use energy. The other configuration is made of six parts – or a hexamer configuration – and serves as a “standby” mode for the protein. “As the name suggests, morphlock-1 essentially locks the hexamer configuration into place, preventing its protein subunits from reconfiguring into the active assembly,” says lead investigator Eileen Jaffe, Ph.D, a Senior Member of Fox Chase. “Targeting morpheeins in their inactive assemblies provides an entirely new approach to drug discovery.” While their study was performed using a pea plant-version of PBGS, the researchers have reason to believe the principle could apply to bacterial versions of PBGS as well. “Using morphlock-1 as a base, we are seeking to fine tune the molecule so that it blocks just the bacterial version of the PBGS enzyme, ” Jaffe says. “Because PBGS is so crucial for life, the part of the enzyme where chemistry happens is highly conserved through evolution,” Jaffe says, meaning that an all-around PBGS-inhibiting drug would harm bacteria, peas and people alike. The area where the potential drug binds to the hexamer form of the protein, however, has been found to differ among species, depending how far the organisms have evolved from each other. When PBGS is in its inactive hexamer form, there is a small cavity on the surface of the assembled complex. Using computer docking techniques, Jaffe and her Fox Chase colleagues identified a suite of small molecules predicted to bind to this cavity. The researchers then bought and tested a selection of these molecules in the lab to see if any of them stabilized the pea PBGS in its hexamer assembly. One inhibitor in particular, given the name morphlock-1, potently drove the formation of the hexamer in pea PBGS, but not in that of humans, fruit flies, or the infectious bacteria Pseudomonas aeruginosa, or Vibrio cholerae, the latter of which causes cholera. Morphlock-1 is a potent inhibitor of pea PBGS, but not of the PBGS from these other organisms. Jaffe coined the term “morpheein” in 2005 after a study of the structure of PBGS revealed its shape-shifting tendencies. While initially met with skepticism because the existence of morpheeins contradicts some classic concepts about protein structure and function, subsequent studies have reinforced that PBGS (and perhaps other proteins) exhibits this behavior. According to Jaffe, this study is the first to make use of alternate morpheein shapes as a potential strategy for drug discovery, in general, particularly for antibiotics. “Multi-drug resistance drives the need for developing new antibiotics,” Jaffe says. “Since drugs that stabilize the inactive PBGS hexamer need not be chemically similar to each other, it will be difficult for the bacterium to develop complete resistance to a cocktail of such compounds.” Source: Fox Chase Cancer Center Josh says: This is a really novel approach. I think in the future, a lot of our drugs, such as antibiotics, will be designed in this way. It makes them much more affective, and it would probably take bacteria longer to evolve resistance to them. Furthermore, it would allow us to quickly develop new antibiotics as resistance becomes more widespread. |
| DDC Announces Launch of Advisor Web Tool [The DNA Testing Blog] Posted: 20 Jun 2008 03:48 PM CDT DNA Diagnostics Center is pleased to announce the recent launch of its Personal DNA Test Advisor on its homepage. The tool is designed to help potential clients find the right DNA test for their situation, and provide information on each type of test so users can determine which type of test they need. "The goal for [...] |
| Mendeley: Discover Research [ScienceRoll] Posted: 20 Jun 2008 03:12 PM CDT I have a constantly updated list of community sites created for researchers and physicians. Here is a new addition, Mendeley that aims to become a useful community where you can manage, share and discover research.
This video describes how the site works: We will see which one will be the winner.  |
| New review of NGS technolgies and ancient DNA research [Next Generation Sequencing] Posted: 20 Jun 2008 02:59 PM CDT I once had the pleasure of visiting David Lambert, an expert and pioneer in ancient DNA studies. With Dave I had some of the most interesting and thought provoking discussion about biology, that I have ever had. Now Dave and his colleagues have a new review out in TREE on New developments in ancient genomics [...] |
| Getting Our Congressional Candidates to Talk About Science [adaptivecomplexity's column] Posted: 20 Jun 2008 02:26 PM CDT Scientists and Engineers for America have put up seven questions that they suggest asking Congressional candidates (see their website for the full questions; the following is abridged): 1. What policies would you support to ensure that America remains the world leader in innovation? 2. What is your position on the following measures that have been proposed to address global climate change—a cap-and-trade system, a carbon tax, increased fuel-economy standards, and research? Are there other policies you would support? 3. What policies would you support to meet the demand for energy while ensuring an economically and environmentally sustainable future? 4. What role do you think the federal government should play in preparing K-12 students for the science and technology driven 21st Century? 5. What policies would you support to meet demand for water resources? 6. Given that the next Congress will likely face spending constraints, what priority would you give to investment in basic research in upcoming budgets? 7. How do you see science, research, and technology contributing to improved health and quality of life? Most of these questions are good ones, and each of these subject areas is something your Congressional representative should have a thought-out answer for. It is glaringly obvious though, that these questions scrupulously avoid areas of science that have been caught up in the culture wars, like stem cell research and evolution vs. intelligent design. |
| Evidence-based web 2.0 [ScienceRoll] Posted: 20 Jun 2008 01:51 PM CDT Dean Giustini is the real pioneer of medicine 2.0. As you may know medicine 2.0 is about the impact of web 2.0 on medicine and healthcare. Giustini is the administrator of UBC HealthLib-Wiki - A Knowledge-Base for Health Librarians. That is a unique resource dedicated to this special field of medicine. He has recently defined what evidence-based web 2.0 means:
Check these interesting pages out:
I’m trying to create a similarly useful collection at Medicine 2.0.  |
| Yep, it's a dog eat dog world, alright [Discovering Biology in a Digital World] Posted: 20 Jun 2008 01:15 PM CDT  "Let this sleepin' dog lie, son. Dog-gone it, I'm dog tired. I'm tired of leading the dog's life and fightin' likes cats and dogs against cats and dogs, a young pup's doggin' my trail tryin' to become top dog. I'm going to the dogs in a dog eat dog world, son. I... I'm so far over the hill... I'm on the bottom of the other side. "- Wylie Burp from Fievel Goes West
|
| Updates on the CCRG and C-51 [Bayblab] Posted: 20 Jun 2008 12:38 PM CDT We received the following email from a reader, updating us on some goings-on with respect to Bill O'Neill and the Canadian Cancer Research Group as well as Bill C-51. [Links added] Hi, Bayblab . . .Quickly checking some of those sites, Project False Hope seems like a nice idea, but as our reader points out, is largely toothless. What it does have is a dummy natural product site and quiz to help identify health fraud, but little else (and no mention of particular companies or groups that should be avoided). I wasn't able to find specific details about the proposed changes to C-51, but it sounds like the revised bill will create a separate category for natural health products (the current version has them grouped with prescription drugs as 'therapeutic products') which takes acknowledges "that natural health products are generally of lower risk and that their long history of use has some value." [Globe and Mail] I'll be interested to see if the proposed changes alter any of C-51's language dealing with standards of evidence and marketing for natural products which I thought was one of the major benefits of the original bill. |
| Thinking about biological resources [business|bytes|genes|molecules] Posted: 20 Jun 2008 12:20 PM CDT
I’m going to focus on one question. Centralization and data quality. In general, to deposit something in a central repository, you should have the following criteria
But those are not necessarily sufficient. Part of the problem with NCBI is the UI, which I’ve never liked, too many databases and somewhat confusing interoperability. There is also a couple of fundamental philosophical questions
The problem with the latter is that they tend not to work in practice, although there are examples. Distribution is more important than centralization. The question show not be “how do we store our data”, but “how do we enable scientists to do more with all the data that we have”. The “how do we store” then becomes one of many questions that need to be asked to answer the primary question. The quality and reliability of biological data is paramount. Being able to access it, both via search UIs and web services APIs is critical. We tend to think of these problems primarily from the scientists point of view, which we need to continue to do so, although I am not sure how well we are doing. However, in developing and organizing these resources, I feel we need to bring in some gurus in distributed data and architectures. The data and information lying with NCBI, EBI, etc has the potential to impact our future, and we are going to continue to see data volumes grow exponentially. Which is why I wonder and hope, if there will ever be something like Wikipedia, or perhaps a Google (not just a search engine, but an organization that makes organizing and indexing life science information a mission statement) that will revolutionize what we do with all the genomes and other associated data at our disposal. Technorati Tags: GenBank, Biological Information, Web Services, APIs, Data Management |
| Tree of Life Runners Web Notes 6-20-2008 [The Tree of Life] Posted: 20 Jun 2008 09:56 AM CDT Just some little notes here in lieu of more detailed posts.
|
| Females in families of homosexual men have higher fertility [Yann Klimentidis' Weblog] Posted: 20 Jun 2008 09:51 AM CDT Sexually Antagonistic Selection in Human Male Homosexuality Andrea Camperio Ciani, Paolo Cermelli, Giovanni Zanzotto PLoS ONE 3(6): e2282 Abstract Several lines of evidence indicate the existence of genetic factors influencing male homosexuality and bisexuality. In spite of its relatively low frequency, the stable permanence in all human populations of this apparently detrimental trait constitutes a puzzling 'Darwinian paradox'. Furthermore, several studies have pointed out relevant asymmetries in the distribution of both male homosexuality and of female fecundity in the parental lines of homosexual vs. heterosexual males. A number of hypotheses have attempted to give an evolutionary explanation for the long-standing persistence of this trait, and for its asymmetric distribution in family lines; however a satisfactory understanding of the population genetics of male homosexuality is lacking at present. We perform a systematic mathematical analysis of the propagation and equilibrium of the putative genetic factors for male homosexuality in the population, based on the selection equation for one or two diallelic loci and Bayesian statistics for pedigree investigation. We show that only the two-locus genetic model with at least one locus on the X chromosome, and in which gene expression is sexually antagonistic (increasing female fitness but decreasing male fitness), accounts for all known empirical data. Our results help clarify the basic evolutionary dynamics of male homosexuality, establishing this as a clearly ascertained sexually antagonistic human trait. |
| Biologists vs. the Age of Information [Discovering Biology in a Digital World] Posted: 20 Jun 2008 08:51 AM CDT It's pretty common these days to pick up an issue of Science or Nature and see people ranting about GenBank (1). Many of the rants are triggered, at least in part, by a wide-spread misunderstanding of what GenBank is and how it works. Perhaps this can be solved through education, but I don't think that's likely. People from the NCBI can explain over and over again that some of the sequence databases in GenBank are meant to be an archival resource (2), and define the term "archive," but that's not going to help. Confusion about database content and oversight is widespread in this community with good reason. Read the rest of this post... | Read the comments on this post... |
| Folding can be fun [Bitesize Bio] Posted: 20 Jun 2008 07:49 AM CDT
If you’ve been interested in the Folding at Home project, but wanted to “play” (pun intended) a much more active role then the wait is over. Join Fold It!
I came across this interesting project from the HHMI Research News arcticle.The game is a project developed by the Baker Lab.You can find more details about the game here. Have fun folding. |
| Forterra: Medical Simulations and Hospital Training [ScienceRoll] Posted: 20 Jun 2008 07:40 AM CDT I’ve written dozens of posts about the medical educational opportunities of Second Life, the virtual world. But we have to make it clear: even if we have great buildings in Second Life, Linden Lab might sell the whole virtual world anytime. Many of the residents think they have no guarantee for their facilities. Second, we focus on case studies and brainstorming during our medical exercices and not on practical examinations due to the basic features of Second Life. Forterra is trying to solve this problem from a different aspect:
What do they provide?
Here is an example of hospital training with the Forterra system: Second Life is for free but we have to deal with limited possibilities while Forterra is, of course, not for free, but totally secure.  |
| I hate it when that happens. [T Ryan Gregory's column] Posted: 20 Jun 2008 07:11 AM CDT I sometimes joke that you should never read your own papers once they are published, because you will undoubtedly find something you wish you had written differently. |
| Learning versus evolution in microbes. [T Ryan Gregory's column] Posted: 20 Jun 2008 06:55 AM CDT One of my pet peeves is the common description in the media of bacteria "learning" to "outsmart" antibiotics. As anyone with a basic comprehension of evolution knows, learning has nothing to do with it. Learning is what happens during the lifetime of an individual, and it occurs in direct response to some information that the individual encounters. When bacteria become resistant to antibiotics, it is not by learning. The individual bacteria do not sense the antibiotic and change to become resistant. Rather, individual bacteria in a population that happen to be resistant because of some genetic difference (or in whom a mutation conferring resistance arises by chance or through gene transfer from another population) will survive and reproduce more effectively than individuals lacking the genetic characteristic that confers resistance. Over many generations of this process, the gene providing resistance to the antibiotic will be found in the majority of bacteria -- not because it "spreads" and not because individual bacteria develop resistance, but because the bacteria that are the most abundant in the population after many generations are obviously the descendants of the ancestors that left the most offspring, namely those who survived the antibiotics. |
| DNA Excerpt: Bringing Home the Birkin [Eye on DNA] Posted: 20 Jun 2008 03:09 AM CDT
Update: For more, see Christina’s book review at eBeautyDaily. |
| The adventure gene [Genetic Future] Posted: 20 Jun 2008 02:32 AM CDT 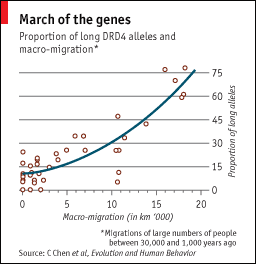 Last week I mentioned a study suggesting that a genetic variant associated with attention deficit disorder (ADD) is beneficial for individuals living in unsettled, nomadic groups but detrimental to those in modern sedentary societies. Last week I mentioned a study suggesting that a genetic variant associated with attention deficit disorder (ADD) is beneficial for individuals living in unsettled, nomadic groups but detrimental to those in modern sedentary societies.The Economist has an interesting article on the study that includes the figure on the left, which is based on data from a 1999 study of 2,320 individuals from 39 populations. Basically, it shows the distance that each population has migrated over the last 1-30,000 years on the x-axis, and the frequency of DRD4 "long alleles" (version of the gene closely related to the ADD-associated version) on the y-axis. There is an intriguing correlation, suggesting that the "novelty-seeking" behaviour associated with ADD may have extended into a desire to explore new territories. Razib has been discussing DRD4 quite a bit recently over on GNXP, including a potential gene-society interaction influencing political beliefs. (Thanks to Simon, who needs to start his own blog ASAP.) |
| New PMed Blog: The Experimental Man [Think Gene] Posted: 19 Jun 2008 11:44 PM CDT David Duncan has started a new website, The Experimental Man, to report just how much one can learn about oneself with modern pmed testing… by taking all the tests himself. From his introduction blog post:
A book is due in early 2009. We’re looking forward to it! |
| Shortest Response: Why do scientific theories work? [Think Gene] Posted: 19 Jun 2008 11:08 PM CDT John Wilkins of Evolving Thoughts conjects in response to The Inherihent Problem with Scientific Theories:
I can compose an infinite responses. I write this one. Have I produced useful information? Yes. Can I compose viable responses infinitely? Yes —that’s called “blogging.” Buy Google stock. Science produces useful information, but the process of science has no end, nor does a single perfect theory exist. Yes, this is semantics, but in the past (until about the 1930’s) scholars believed that the universe could be fully understood and a perfect model could be known. This is no longer the contemporary belief. |
| Researchers find an evolutionarily preserved signature in the primate brain [Think Gene] Posted: 19 Jun 2008 10:07 PM CDT Researchers from Uppsala University, Karolinska Institute, and the University of Chicago, have determined that there are hundreds of biological differences between the sexes when it comes to gene expression in the cerebral cortex of humans and other primates. These findings, published June 20th in the open-access journal PLoS Genetics, indicate that some of these differences arose a very long time ago and have been preserved through the evolution of primates. These conserved differences constitute a signature of sex differences in the brain. More obvious gender differences have been preserved throughout primate evolution; examples include average body size and weight, and genitalia design. This novel study focuses on gene expression within the cerebral cortex – that area of the brain that is involved in such complex functions in humans and other primates as memory, attentiveness, thought processes, and language. The researchers measured gene expression in the brains of male and female primates from three species: humans, macaques, and marmosets. To measure activity of specific genes, the products of genes (RNA) obtained from the brain of each animal were hybridized to microarrays containing thousands of DNA clones coding for thousands of genes. The authors also investigated DNA sequence differences among primates for genes showing different levels of expression between the sexes. “Knowledge about gender differences is important for many reasons. For example, this information may be used in the future to calculate medical dosages, as well as for other treatments of diseases or damage to the brain,” says Professor Elena Jazin of Uppsala University. Lead author Björn Reinius notes that the study does not determine whether these differences in gene expression are in any way functionally significant. Such questions remain to be answered by future studies. Source: Public Library of Science Josh says: I’d be curious to see how these differences compare to the expression levels in homosexual males and females. |
| Posted: 19 Jun 2008 09:28 PM CDT A collaboration of scientists from Iceland and the United States has used Iceland’s genealogical database (by deCODE genetics) to trace the ancestors of patients suffering from hereditary cystatin C amyloid angiopathy (HCCAA). Analysis shows that the deadly mutation in the cystatin C gene, L68Q, derives from a common ancestor born roughly 18 generations ago, around 1550AD. Details are published June 20th in the open-access journal PLoS Genetics. This dominantly inherited disease, which is due to a mutation in cystatin C (L68Q), strikes young adults with healthy blood pressure. The disease results in death from repeated brain haemorrhages, on average by the age of 30. The origin of the mutation causing HCCAA was previously unknown, but using DNA haplotype analysis the scientists have shed light on the history of this autosomal dominant disease that has high penetrance in contemporary Icelanders. The scientists found that 200 years ago, obligate carriers of the mutation lived a normal life span compared to the control population (their spouses). In carriers born around 1820, however, a trend of shortening life span began, resulting in an average life span of only 30 years in people born around 1900. This 30-year lifespan has stayed constant since then in both men and women. At the same time, a matrilinear effect appeared whereby those who inherited the mutation from the mother died earlier. For carriers born after 1900, the difference is a loss of 9.4 years for those who inherited the mutation from their mothers rather than their fathers. Based on this information, the authors propose that the traditional diet of the nation (which in the past consisted largely of whey-preserved offal as well as meat, dried fish, and butter) “protected” the mutation carriers for almost 300 years until the Icelandic diet changed early in the early 19th century, exemplified by drastic increases in imported carbohydrates and salt. This finding has implications for studies of Alzheimer’s disease as cerebral amyloid angiopathy (CAA) is almost universally found in Alzheimer’s patients and normal cystatin C protein is one of the proteins found in amyloid in brains of Alzheimer’s patients. Studies are underway to try to elucidate the risk factors with the hope of providing a preventive stategy for cystatin L68Q carriers. Source: Public Library of Science Josh says: This is amazing. It almost makes me wish that the United States and other countries had a database like Iceland does, except I don’t really trust the US government. Regardless, as the costs for sequencing decrease, we should start to see more discoveries like this. |
| Demonstrate evolutionary innovation, win $20,000 [Genetic Future] Posted: 19 Jun 2008 08:30 PM CDT  The InnoCentive website is a kind of ideas marketplace where "Seekers" pose detailed descriptions of theoretical or technical problems in their field and offer financial incentives for "Solvers" to figure them out. The InnoCentive website is a kind of ideas marketplace where "Seekers" pose detailed descriptions of theoretical or technical problems in their field and offer financial incentives for "Solvers" to figure them out.One of this week's challenges would be of interest to all the theoretical and experimental evolutionary biologists out there, especially given the $20,000 reward money on offer: During the evolution of life on Earth new biological features have emerged in a process that continues without reaching an obvious maximum level of organized complexity, even now. The Seeker is interested to know if this apparently open-ended evolutionary innovation would be possible in a quarantined system. If so, can it be demonstrated? The goal of this theoretical challenge is to come up with an acceptable demonstration design to provide a positive result.You'll need to register as a Solver to read the details of the challenge (which requires agreeing to not disclose the information therein). The Seeker has also set up a Google Groups page to discuss the issue, where someone has already suggested that this process was well-demonstrated a fortnight ago by Richard Lenski's group. Of course, it all depends on your definition of "innovation" - and I hope I'm not crossing any legal boundaries by saying that the Seeker's definition in the detailed challenge is far from satisfying. |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |










No comments:
Post a Comment