The DNA Network |
| How she really does it: A Radio Talk Show for Women [Tomorrow's Table] Posted: 27 Jun 2008 06:55 PM CDT Today I had the pleasure of conversing with Koren Motekaitis who is host of a public radio show here in Davis called A Radio Talk Show for Women "How She Really Does It" covers a variety of topics, including balancing work and family, financial issues, career, building businesses, parenting, education, religion, and youth sports. This talk show is meant to inspire, empower, entertain and be a resource for women. We discussed organic farming, genetics and the future of food as well as other important topics such as hanging your clothes out to dry. To hear the interview, please check out the podcast |
| The Evolutionary History of Birds [evolgen] Posted: 27 Jun 2008 05:30 PM CDT Phylogeny Friday -- 27 June 2008 I haven't done a Phylogeny Friday in about a month, but a recent paper reporting a "phylogenomic study of birds" was worth mentioning (doi:10.1126/science.1157704). Now, this isn't phylogenomics as Jonathan Eisen defined it. The bird evolution paper describes building a tree using lots of molecular markers. I don't have much to say about the new bird phylogeny (I'll let the expert handle the details), but I wanted to post one of the trees for Phylogeny Friday. Here is one of the phylogenies they present, attempting to reconcile their results with those that had been previously published: The three columns at the tips indicate three previously published taxonomic classifications. Those with black text are monophyletic taxa in this new study, while those with white text are paraphyletic. The different colored lineages indicate various clades that are supported by this new study (e.g., green indicates land birds, blue are water birds, etc.). Eisen. 1998. Phylogenomics: Improving Functional Predictions for Uncharacterized Genes by Evolutionary Analysis. Genome Res. 8: 163-167 [link] Hackett et al. 2008. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science 320:1763-1768 doi:10.1126/science.1157704 Read the comments on this post... |
| Another Disease (almost) Eradicated [Epidemix] Posted: 27 Jun 2008 04:00 PM CDT
Quick background: Rinderpest is a viral disease that afflicts livestock, mainly cattle. It is brutal, often killing a third of a herd. A century ago, it spread throughout Asia, Africa, and Europe, but various efforts, culminating in a sustained international campaign, begun in 1994, has driven it to isolated patches in Africa. Lately, it’s been confined to Kenya, and now may be even gone from there. Just because it’s a bovine disease, though, doesn’t mean it doesn’t have human impact. Consider this passage from the Science story (subscription required), describing an outbreak in South African in 1897 that killed about 90% of the cattle population, as well as other livestock and local game:
More recent outbreaks have likewise proven devasting to cattle and human populations alike; a particularly virulent outbreak in Sudan in the late 1980s killed 80% of calves, and with cow milk unavailable, human children began to starve, resulting in a horrible famine. Another example of how we’re just a part of one big ecosystem. Eradication is on target for 2010. Can’t wait to toast this one. |
| The Public Health Case for Direct-to-Consumer Personal Genomics [Epidemix] Posted: 27 Jun 2008 02:27 PM CDT OK - A couple more thoughts on this move by health departments in California and New York to regulate personal genomics. I’ve made my quasi-libertarian case that this is my information and shouldn’t be mediated by an under-informed (and possibly antagonistic) physician gatekeeper. And I’ll leave the companies to make their own case on the issue of lab oversight. But now let me make an argument on public health grounds - the home turf, after all, of these state agencies. To my mind, their actions will directly contravene their own mandate, and will have the result of reducing the public’s health. The California DPH says it’s acting to “protect consumers.” As Wired Science’s Alexis Madrigal ferreted out, in a nifty bit of reporting, CDPH’s Karen Nickel said in a June 13 meeting that the state’s primary concern is that personal genomics companies are creating the “worried well” - citizens who stumbled into a level of knowledge about their genome that they were unprepared for, and now have may be fretting in a a way detrimental to their health & well being. Put aside the fact that, as a public health matter, the “worried well” is a supremely thin basis for action (what pray tell is the prevelence of “worried well” in California? The incidence? The relative risk of learning one’s genome? What sort of epidemiological studies have been performed to measure this population?). And put aside the fact that, as others have noted, the customers of 23andMe and Navigenics and other personal genomics companies are, in demographic terms, probably the *least* likely to be categorized as “uninformed” or naive. These are early adopters, they’re paying lots of money (opting-in), and are probably far more prepared to reckon with genomic information than the typical citizen. But put that all aside. My argument is simply that by restricting personal genomics to a physician-vetted service, these state public health departments would be eviscerating the actual public-health utility from genomics. The whole *point* of learning one’s genomic predispositions is as a predictive and preventative tool. Learn early, so as to change our behaviors, intervene early, and either skirt or reduce the prospects of disease. This is a *long term* tool. But by regulating the service, these state health departments would severely impinge the opportunity to make the largest public health impact, in two specific regards: 1) Public health, by definition, is about populations, not individuals, and Nickels makes a quasi-population argument when she identifies this group of “worried well.” OK, let’s take a stab at quantifying this. The worried well would be some fraction of personal-genomics customers; let’s give Nickels a big gimme and say that 20% of customers would somehow overreact in a way that’s detrimental to their health (I’m of course making that up, and it’s almost absurdly high, but let’s go with it). Stress would be the most obvious detriment, but it could be something like taking unnecessary medication or supplements, etc. Now consider what percentage of personal-genomics customers actually engage in their genomic information in a way that’s *beneficial* to their health, as intended. These people pay their money, get their results, spot their risks, and change their lives, often in small ways or rarely in big ways. Let’s low-ball it and say that 60% of all customers act on their results (paying $1,000 or $2,500 is actually a significant motivator, but let’s assume 40% just ignore the results altogether). And of course not all results would be positive; some would be null. So let’s take another slice and say some fraction of the 60% actually measurably benefit, either in peace of mind or in some slight amelioration of diet and exercise or doctor’s visits. Let’s say half of the 60% - so 30%. Even at that low number, that is very clearly a net positive - the public’s health at large has been improved. Of course, I’m making up these figures, and really it’s probably impossible to measure (though I bet 23andMe and Navigenics are crafting customer surveys to help fill in this picture). But really, by any informed assessment, the net potential for improving the public’s health far outweighs the possible detrimental effects of “worried well.” Just as vaccines and even exercise already have their detrimental side effects, sure, so will personal genomics. But if you’re tasked with improving the public’s health, as these agencies are, why not consider the benefits as well as the risks? 2) So genomics are useful as a predictive tool, they give us a peek into our longterm health prospects and an opportunity to intervene and improve those prospects. The fact that consumers in California can, for the moment, engage in that information now at their own behest means they are getting the information when they want it, which is by definition as early as possible. So what’s the logical consequence of forcing a physician into the picture as a middleman? Well, it’s a pretty good guess that it’ll delay people from getting the information. Put physician-phobias and reluctance to schedule a visit and all sorts of other procrastinations together, and I think this would result in less and later genotyping; both a significant delay in when this information reaches people - as well as a significant reduction in the number of people who actually bother to jump through these extra hoops. The net result, again: a squandered opportunity to impact the public’s health. I would be the first to acknowledge that the actual science is fairly raw here; we’re in the early days of using our genomes for actual health decisions. But that’s the point: better to get familiar with the information now, when it’s fairly low-impact, and work out the kinks than to wait for the science to somehow emerge fully-formed and neatly packaged. Because if we’re waiting on the physician community for that day, it’ll never come. But assuming the public health department acknowedges that genetics *does* have some utility for our health, then I’d remind them that a fundamental principle of public health is awareness - give citizens information earlier so that they can avoid putting themselves at risk. That principle drives public health’s actions against smoking, infectious disease, sexually transmitted disease, natural disasters, and so many other threats. Likewise, it drives their actions on positive behaviors like proper nutrition and exercise. So why, in the case of genomics, should the same principle not apply? Why, in this case, do state health departments think the public should be prevented from learning about their risks? |
| Posted: 27 Jun 2008 02:16 PM CDT Following the debate surrounding the FDA's 2005 approval of BiDiL – a drug to be marketed to treat African-Americans at risk for heart failure – David e. Winickoff and Osagie K. Obasogie propose regulatory policy for future race-specific drug development. Writing in a letter published in Trends in Pharmacological Sciences [Race-specific drugs: regulatory trends and public policy. 2008 Jun;29(6):277-9. Epub 2008 Apr 29 | PMID: 18453000 - CiteULike (excerpt)], the authors argue: "race-specific indications should be rejected unless clinical trials can demonstrate convincingly that the drugs are both better than existing treatments for a specific group and no better than existing treatments for non-specified groups". They conclude that these enhanced regulations might help to reduce health disparities while protecting groups from market exploitation: "Race can be used as a proxy for the group most likely to benefit from a drug as long as the effect is not to deny others valid treatments". In other words, "Pharmaceutical science and biomedicine most certainly should not be colorblind. But they also must not be 'color-struck'". Unlike one's genetic information, racial identity is a social-construct – so, using race as a proxy for individuals with common genetic characteristics is a messy and controversial process. In this case, would it make sense at all to say: genome-specific "indications should be rejected unless clinical trials can demonstrate convincingly that the drugs are both better than existing treatments for a specific group and no better than existing treatments for non-specified groups"? Would such a standard be useful or does it merely re-state the obvious? – J.O. |
| Interview with Jenny Graves in PLoS Genetics [evolgen] Posted: 27 Jun 2008 02:00 PM CDT PLoS Genetics has published an interview with Jenny Graves. Graves is one of the leaders in monotreme and marsupial genetics, and has been involved in some of the recent mammalian genome projects, including the platypus genome project (doi:10.1038/nature06936). She is also an expert in the evolution of mammalian sex chromosomes and sex determining genes. However, I'd like to point to a quote in PLoS Genetics' interview of Graves that deals with science education:
The entire interview is interesting, and it touches on many of the highlights (and lowlights) of Graves' career. Read the comments on this post... |
| Posted: 27 Jun 2008 01:15 PM CDT OK, I am getting worried. As I wrote a few weeks ago, there is a new movie coming out supposedly next year called "The Tree of Life" starring Brad Pitt, Sean Penn and others. Well, the movie sounds like it could be good. But though this may sound like a cool thing to those of you out there who study the Tree of Life in some way, we are at risk here. There will soon be 100s of blogs, web pages, news stories, etc. writing about the Tree of Life movie. And uses of the Tree of Life by evolutionary biologists will lose their google rankings. The term "The Tree of Life" is at risk of a form of extinction. So what are we to do? I am calling on all evolutionary biologists and bloggers and biologists in general to to link to other sites that use the term as much as possible. For example, you can add a link to my blog, which is after all called "The Tree of Life." So please, add links to some Tree of Life pages. And start using the term yourself as much as possible. Here are some places you should link to
Please post additional links in the comments here and please help save "The tree of life" |
| Cancer Carnival Call for Submissions [Bayblab] Posted: 27 Jun 2008 12:54 PM CDT  The 11th Edition of the Cancer Research Blog Carnival is coming in one week. This July 4th Holiday edition will be hosted here at the Bayblab. The 11th Edition of the Cancer Research Blog Carnival is coming in one week. This July 4th Holiday edition will be hosted here at the Bayblab. UPDATE: Submissions can now be made at blogcarnival.com, here. |
| Posted: 27 Jun 2008 10:22 AM CDT Hakia is a semantic web search engine, so what you might think. The difference is hakia targets legal, financial and Medical web searches. Thye have even licensed its technology to a startup company that summarized information for government and pharmaceutical companies There is more starting from April 2008 using Hakia you can search Pubmed. Adding more than 10 million abstracts from PubMed to their index and setting up a new site dedicated to searching this content at pubmed.hakia.com. The PubMed content will also be visible in search results on the hakia medical search site and via the main hakia search page For exmaple my query of microarray software Pubmed returned 1646 results and Hakia 15 some times having too much is not good idea, especially when you are looking for specific text. It doesnt even stop there, the result also showed me Still not convinced of the merits take a look at the comparison of the search results on new regulatory problems affecting personal genomics companies, in google and Hakia Semantic search startup Hakia June 19 made its Syndication Web Services APIs available to search engine marketing or publishing businesses and others that want to provide their users with better ways of finding information. The XML search option and feed syndication search servicces are certinaly good Hakia is offering 30,000 free (and advertising-free) searches per day to early adopters until the partners’ quota is filled. You can also generate a custom seach box that can be added to your website much like the custom google search box . and yes there is competition to Powerset has launched its first product May 12, a search engine focused on finding articles on Wikipedia. And there is roumor that Microsoft will acquire Powerset Semantic Search engine very soon for nothing less but USD 100M May be because Yahoo is already offering Semantic webs earch using through its SearchMonkey platform But there is more than what meets the eye. Remember Interestingly in 2007 June Oracle staked its claim to leadership in the enterprise side of the emerging semantic Web space, saying that more than 100 commercial and open-source applications are using its version of the technology. Oracle do offer search called Oracle Secure Enterprise Search. Take a look at the Oracle Semantic Technology Centre Microsoft can also say to be making up for the loss of TripleHop to Oracle, There is still more surprises Evri.com a Paul Allen backed semantic search engine has launched a Beta release in June 24. Other similar service providers includes TextDigger, Radar Networks Peer39.com that offers semantic technology in advertising, Gloofi.com Twine, then there is Spock calling itself a vertical search engine for people, usign top-down semantic search. In that sense we can call Google Maps to be using semantic technology though it cant be called a thorough breed in this field
|
| Geeky DNA T-Shirt: XX Chromosomes [Eye on DNA] Posted: 27 Jun 2008 09:52 AM CDT |
| Great time on Day 1 [business|bytes|genes|molecules] Posted: 27 Jun 2008 09:06 AM CDT The first day on Day 1 of the New Communication Channels for Biology workshop was great. Lots of interesting talks and Q&A sessions at a very good venue. More after I return to Seattle, but it is good to see scientists discuss wikis, blogs, video, open science, etc openly, without too much rhetoric. There are also many people really leveraging wikis and other platforms for real science, and doing a pretty good job of thinking about APIs and making sure their resources exist on the programmable web. Also got a chance to plug FriendFeed :). |
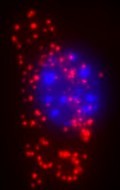
| Lighting Up Genetic Disease [Sciencebase Science Blog] Posted: 27 Jun 2008 07:00 AM CDT
There are some genetic diseases, however, that are apparently caused by nothing more than a single mutation in the human genome. A single DNA base out of the many thousands on a person’s genetic material, if swapped for another the wrong base means the production of the protein associated with that gene goes awry. For example, sickle cell disease is caused by a point mutation in the gene for beta-haemoglobin. The mutation causes the amino acid valine to be used in place of glutamic acid at one position in the haemoglobin. This faulty protein cannot fold into its perfect active form, which in turn leads to a cascade of effects, that result ultimately in faulty red blood cells and their associated health problems for sufferers. There are many other diseases associated with single point mutations, including achondroplasia, characterised by dwarfism, in one sense, cystic fibrosis, although different single mutations may be involved, and hereditary hemochromatosis, an iron overload disease. “To be able to study and diagnose such diseases with limited material from patients, there is a need for methods to detect point mutations in situ,” explains Carolina Wählby of the Department of Genetics and Pat Centre for Image Analysis, at Uppsala University, Sweden, and her colleagues Patrick Karlsson, Sara Henriksson, Chatarina Larsson, Mats Nilsson, and Ewert Bengtsson. Writing in the inaugural issue of the International Journal of Signal and Imaging Systems Engineering (2008, 1, 11-17), they pointed out that this is a problem that can be couched in terms of “finding cells, finding molecules, and finding patterns”. The researchers explain that the molecular labelling techniques used by biologists in research into genetic diseases often just produce bright spots of light on an image of the sample. Usually, these bright spots, or signals, are formed by selective reactions that tag specific molecules of interest with fluorescent markers. Fluorescence microscopy is then used to take a closer look. However, signals representing different types of molecules may be randomly distributed in the cells of a sample or may show systematic patterns. Such patterns may hint at specific, non-random localisations and functions of those molecules within the cell. The team suggests that the key to interpreting any patterns of bright spots relies on slicing up the image quickly, applying signal detection, and finally, analysing for patterns. This is not a trivial matter. One solution to this non-trivial problem could lie in employing data mining tools, but rather than extracting useful information from large databases, those tools would Biological processes could thus be studied at the level of single molecules, and with sufficient precision to distinguish even closely similar variants of molecules, the researchers say, revealing the intercellular and sub-cellular context of molecules that would otherwise go undetected among myriad other chemicals. The team has demonstrated proof of principle by developing an image-based data mining system that can look for variants in the genetic information found in mitochondria (i.e. mitochondrial DNA, mtDNA). They point out that MtDNA is present in multiple copies in the mitochondria of the cell, is inherited together with cytoplasm during cell replication and provides an excellent system for testing the detection of point mutations. They add that the same approach might also be used to detect infectious organisms or to study tumours. A post from David Bradley Science Writer |
| What it’s like to be a bat [Think Gene] Posted: 26 Jun 2008 11:41 PM CDT Not many people think about what it’s like to be a bat, but for those who do, it’s enlightening and potentially groundbreaking for understanding aspects of the human brain and nervous system. Cynthia Moss, a member of the Neuroscience and Cognitive Science program at the University of Maryland, College Park, Md., is one of few researchers who spend time trying to get into the heads of bats. Her new research suggests there is more to studying bats than figuring out how they process sound to distinguish environments. Partially supported by the National Science Foundation, her research paper appears in the June 18 online edition of the Proceedings of the National Academy of Sciences. “For decades it’s been recognized that a bat’s voice produces sounds that give the bat information about the location of objects,” says Moss. “We’re now recognizing that every time a bat produces a sound there are changes in brain activity that may be important for scene analysis, sensorimotor control and spatial memory and navigation.” The research could help neurobiologists understand mechanisms in the human brain and ultimately benefit human health, but that may not happen for some time as more research is needed. Moss and her colleague, Nachum Ulanovsky from the Weizmann Institute of Science in Israel, reviewed more than 100 studies and determined the brief calls emitted through a bat’s mouth or nostrils and their returning echoes play a pivotal role in motor control and have other behavioral implications. In short, echoes from a bat’s voice cause the bat to turn its head and ears, and give the bat’s brain a description of the scene. The echoes also cue a bat’s memory about its environment so it can safely fly between points. “Our review highlights new research findings suggesting that the bat’s vocal production does more than yield echoes,” says Moss. “We’re learning every time the bat produces a vocalization, there are changes in brain activity that are essential to complex behaviors.” For example, when a bat pursues prey, a moth or some other insect, it computes the 3D location of objects in its environment–a tree, a wall or a lamppost–from information carried by the echoes of high-pitched vocal chirps produced at rates of 2-150 chirps per second. Research shows it actually uses these echoes to remember details of the environment in which it operates, displaying a very sensitive spatial memory component. The vocalizations tell the bat the horizontal and vertical positions of its prey from differences in the arrival time, intensity, and variety of echoes it receives. It estimates target range from the time delay between the outgoing vocalization and returning echo. It also uses its sonar system to assess the size of a target. Finally, when zeroed in, it swoops down on its prey. Researchers are able to draw correlations between how bats and humans process information to perform functions. Both are mammals having the same basic brain organization, which leads to obvious comparisons. According to Moss, bats engage in vocal-motor behaviors to generate signals to probe the environment, while some blind humans produce tongue clicks to generate sounds for echolocation. Both bats and humans engage in so-called “motor behaviors” that shape their perceptions of the world. A bat turns its head, moves its ears, and changes its flight path in response to echo information from the environment. A human moves his eyes or turns his head to augment his perception of auditory and visual signals. “All of these motor behaviors influence the animal’s perception and representation of the environment,” says Moss. “But of interest to us is the idea that these vocal-motor behaviors contribute to environmental perception, memory, and spatial planning far beyond the processing of sound.” “It may be that some of the information we learn from the bat gives us a window into understanding mechanisms of the human brain,” says Moss. “But those outcomes are a little bit down the road.” Source: National Science Foundation Josh says: I’ve always wanted to have echolocation. The mammalian brain is very adaptive, and it can make use of any input, so long as it’s present during neurological development. It’s logical then that bats react to this sense much in the same way we respond to our senses. Andrew says: Humans do use ambient sound to navigate. I’ll try to find the source and post it here. This isn’t what I meant, but here’s a video of some blind kid who might use crude echolocation. |
| Posted: 26 Jun 2008 09:38 PM CDT From over at Wired
Do we need scientific thinking at all anymore? |
| How's that for Genomic Medicine by Press Release? [The Gene Sherpa: Personalized Medicine and You] Posted: 26 Jun 2008 08:48 PM CDT |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |





 Genetic disease is a complicated affair. Scientists have spent years trying to find genetic markers for diseases as diverse as asthma, arthritis and cardiovascular disease. The trouble with such complex diseases is that they are none of them simply a manifestation of a genetic issue. They involve multiple genes, various other factors within the body and, of course, environmental factors outside the body.
Genetic disease is a complicated affair. Scientists have spent years trying to find genetic markers for diseases as diverse as asthma, arthritis and cardiovascular disease. The trouble with such complex diseases is that they are none of them simply a manifestation of a genetic issue. They involve multiple genes, various other factors within the body and, of course, environmental factors outside the body.
No comments:
Post a Comment