Spliced feed for The Science Network |
| Lighting Up Genetic Disease [Sciencebase Science Blog] Posted: 27 Jun 2008 07:00 AM CDT
There are some genetic diseases, however, that are apparently caused by nothing more than a single mutation in the human genome. A single DNA base out of the many thousands on a person’s genetic material, if swapped for another the wrong base means the production of the protein associated with that gene goes awry. For example, sickle cell disease is caused by a point mutation in the gene for beta-haemoglobin. The mutation causes the amino acid valine to be used in place of glutamic acid at one position in the haemoglobin. This faulty protein cannot fold into its perfect active form, which in turn leads to a cascade of effects, that result ultimately in faulty red blood cells and their associated health problems for sufferers. There are many other diseases associated with single point mutations, including achondroplasia, characterised by dwarfism, in one sense, cystic fibrosis, although different single mutations may be involved, and hereditary hemochromatosis, an iron overload disease. “To be able to study and diagnose such diseases with limited material from patients, there is a need for methods to detect point mutations in situ,” explains Carolina Wählby of the Department of Genetics and Pat Centre for Image Analysis, at Uppsala University, Sweden, and her colleagues Patrick Karlsson, Sara Henriksson, Chatarina Larsson, Mats Nilsson, and Ewert Bengtsson. Writing in the inaugural issue of the International Journal of Signal and Imaging Systems Engineering (2008, 1, 11-17), they pointed out that this is a problem that can be couched in terms of “finding cells, finding molecules, and finding patterns”. The researchers explain that the molecular labelling techniques used by biologists in research into genetic diseases often just produce bright spots of light on an image of the sample. Usually, these bright spots, or signals, are formed by selective reactions that tag specific molecules of interest with fluorescent markers. Fluorescence microscopy is then used to take a closer look. However, signals representing different types of molecules may be randomly distributed in the cells of a sample or may show systematic patterns. Such patterns may hint at specific, non-random localisations and functions of those molecules within the cell. The team suggests that the key to interpreting any patterns of bright spots relies on slicing up the image quickly, applying signal detection, and finally, analysing for patterns. This is not a trivial matter. One solution to this non-trivial problem could lie in employing data mining tools, but rather than extracting useful information from large databases, those tools would Biological processes could thus be studied at the level of single molecules, and with sufficient precision to distinguish even closely similar variants of molecules, the researchers say, revealing the intercellular and sub-cellular context of molecules that would otherwise go undetected among myriad other chemicals. The team has demonstrated proof of principle by developing an image-based data mining system that can look for variants in the genetic information found in mitochondria (i.e. mitochondrial DNA, mtDNA). They point out that MtDNA is present in multiple copies in the mitochondria of the cell, is inherited together with cytoplasm during cell replication and provides an excellent system for testing the detection of point mutations. They add that the same approach might also be used to detect infectious organisms or to study tumours. A post from David Bradley Science Writer |
| Life-seeking robot sub to explore Europa someday? [Earth & Sky Podcast] Posted: 27 Jun 2008 04:06 AM CDT A robot sub being tested now in Antarctica might someday be used to find signs of life on other worlds. For example, Jupiter's moon Europa might harbor life swimming in an ocean covered by ice. This posting includes an audio/video/photo media file: Download Now |
| Ark underway. Amphibians only need apply. [Earth & Sky Podcast] Posted: 26 Jun 2008 04:06 AM CDT The Amphibian Ark is a global effort to collect amphibians in zoos, aquariums, and elsewhere. EarthSky asks: why amphibians? This posting includes an audio/video/photo media file: Download Now |
| You are subscribed to email updates from The Science Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The Science Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |
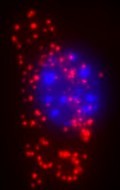 Genetic disease is a complicated affair. Scientists have spent years trying to find genetic markers for diseases as diverse as asthma, arthritis and cardiovascular disease. The trouble with such complex diseases is that they are none of them simply a manifestation of a genetic issue. They involve multiple genes, various other factors within the body and, of course, environmental factors outside the body.
Genetic disease is a complicated affair. Scientists have spent years trying to find genetic markers for diseases as diverse as asthma, arthritis and cardiovascular disease. The trouble with such complex diseases is that they are none of them simply a manifestation of a genetic issue. They involve multiple genes, various other factors within the body and, of course, environmental factors outside the body.
No comments:
Post a Comment