The DNA Network |
| Chris Anderson, you are wrong [business|bytes|genes|molecules] Posted: 25 Jun 2008 08:24 PM CDT Chris Anderson is a man I respect and Wired a magazine I like most of the time. During my chat with Jon Udell, I had bemoaned the gap between science and the general public, and Anderson’s latest article on Wired only serves as a reinforcement of that frustration. In an article entitled The End of Theory: The Data Deluge Makes the Scientific Method Obselete he writes about how the petabyte era is going to change science has one thing very right, and one thing very wrong. He is write more is not more. It is different. But more does not mean the end of the scientific method. I would argue that it allows us to think about new ways to apply the scientific method to try and solve problems. Biology has changed a lot since the 70s, and its not just genetics. The very field has been turned around on its head. What does that mean? It means new methods, new techniques. The human and other genomes, high throughput structure prediction, more data points, might lead to a lot of confusion at first as we try and apply our old methods to new problems and data types, but it also leads to the creation of new techniques, our ability to tap into all this data, to build better models which can help describe what we know. It’s always a moving target. How can we develop better, more refined models that explain all kinds of phenomena. I am a life scientist (although with a physical sciences background). I have spent the better part of the last two years evangelizing “what science can learn from Google”, but I don’t see this as science under threat. I see this as opportunity. How can I (or others who actually still do science) take the new paradigms of computing (by the way, bioinformaticians have been using methods typically used in “collective intelligence” for years), and take biology, which is now very much a digital science, and combine them with our scientific reasoning, our ability to take phenomena and develop models that explain those phenomena and do something meaningful with them. I have seen many computer scientists develop some very elegant theoretical models for biological information, but often without any biological context. Yes scientists need to adopt new techniques, develop new theoretical approaches, even rethink the very basic tenets that they know, but to say the scientific method is dead or approaching the end is sensationalist in the least, and completely uneducated in the extreme. Having to use those words for an article pen by someone whose writing I admire hurts, but science is not a game of hype. It’s hard, and all the easy problems have been solved. Footnote: We are barely tapping into quantum theory to solve life science problems. Most of our methods to look at protein structure and dynamics, to look at drug-protein interactions are either rule-based or Newtonian in nature, with quantum mechanics providing many of the parameters. So lets not put the cart before the horse, at least in the life sciences. I am sure the physicists are trying to develop new theories. That only means physics is very and truly alive. I wonder if people said science was dead when Schrodinger gave the world his equation. |
| Blue Light Retraction [Think Gene] Posted: 25 Jun 2008 08:06 PM CDT We promoted un-reviewed research yesterday regarding a blue light effect on tumors in mice: “Blue light used to harden tooth fillings stunts tumor growth.” This is my fault, and we’ve removed the content. Special thanks to Steven Murphy of “Gene Sherpas” and Matt Mealiffe of “Cancer and Your Genes” for bringing this to our attention. A note: don’t link to dubious content on the Internet (or at least, use a rel=”nofollow”). Otherwise, you help search engines find the bad content and help promote it. Of course, we here at Think Gene want to only promote the best and most interesting bio news, so we will correct mistakes immediately, but many others do not share this belief. |
| Testing, testing, 1, 2, 3 [genomeboy.com] Posted: 25 Jun 2008 07:30 PM CDT As usual, Daniel has an outstanding roundup of the latest developments in the personal genomics kerfuffle (or is it a saga?). If Navigenics wins its argument in California — i.e., interpreting a test is not itself a test and therefore not subject to regulation as such — then will we finally be able to move on to the next existential crisis? In the immortal words of Woody Allen, “I’d call you a sadistic sodomistic necrophile, but that’s beating a dead horse.” |
| Posted: 25 Jun 2008 05:41 PM CDT  Yesterday marked the deadline set by the California Department of Public Health for thirteen genetic testing companies to halt direct-to-consumer marketing and demonstrate their compliance with state regulations. Briefly, the state insists that genetic testing be conducted using specially licensed laboratories, and - more controversially - that all tests be ordered through a clinician (a proposal that stirred up howls of outrage from the human genetics blogosphere). Yesterday marked the deadline set by the California Department of Public Health for thirteen genetic testing companies to halt direct-to-consumer marketing and demonstrate their compliance with state regulations. Briefly, the state insists that genetic testing be conducted using specially licensed laboratories, and - more controversially - that all tests be ordered through a clinician (a proposal that stirred up howls of outrage from the human genetics blogosphere).All thirteen targeted companies are now listed on the health department website. In addition to the "big three" genome-scan companies (23andMe, deCODEme and Navigenics) and the boutique genome-sequencing company Knome (which offers a whole genome sequence for a cool $350,000) there is a bizarre menagerie of less well-known companies. Three of them (Salugen, Sciona and Suracell) are members of the maligned "nutrigenetics" industry, while New Hope Medical Center appears to be a generic unconventional medicine provider that's dabbling in genetics. Gene Essence is a weak attempt by Biomarker Pharmaceuticals to jump on the genome-scan bandwagon. DNATraits, which I've mentioned previously, and Portugal's CGC Genetics both market tests for a variety of rare single-gene diseases. Finally there's HairDX, a highly specialised company that tests polymorphisms associated with hair loss. You can download the letters from the health department's website, although they're hardly riveting reading. I'm surprised to see deCODEme on the list since residents of California (and several other states) are explicitly restricted from using the service's genetic risk calculator. (As an irrelevant aside, I imagine that 23andMe's Linda Avey was slightly annoyed by the consistent misspelling of her last name, and the burly CEO of deCODE would have been less than impressed with being referred to as "Ms. Stefansson"!) The responses to the health department letter have ranged from immediate surrender to preparations for battle. Wired reports that both baldness specialist HairDX and nutrigenetics company Sciona have dropped out of the California market rather than face the legal wrath of the CDPH; SeqWright, which offers a pretty half-hearted genome scan service, packed its bags without even receiving a letter. In contrast, 23andMe and Navigenics - which both presumably have the financial backing to sustain a legal wrangle - are planning to stay. Wired reports on a statement released to them by San Francisco-based 23andMe, which clearly has no intention of losing a foothold in its home state: We believe we are in compliance with California law and are continuing to operate in California at this time. Our testing is conducted in an independent CLIA-certified laboratory and we utilize the services of a California licensed physician. However, we would like to have continued discussions with the Department regarding the appropriate regulation of this unique industry.23andMe only recently moved to a lab that was certified under the Clinical Laboratories Improvement Act of 1988 (CLIA), presumably as an attempt to head off this sort of regulatory crack-down. So basically, they're arguing that they're not in breach of the rules and will continue to test California residents until the law changes. Navigenics, meanwhile, has sent a detailed defence of their position to the health department - and it starts with a much more audacious argument: In a letter sent to the Health Department obtained by Wired.com, the company argues that it does not actually perform genetic tests, and therefore should not be regulated as a clinical laboratory under California state law.Because the company out-sources the actual genome scan to a certified lab run by Affymetrix, it never actually touches the DNA - it only handles the information derived from that DNA. Wired comments: Navigenics is arguing that once the state-licensed lab turns a biological sample into digital data, DNA is no longer within the purview of health department laboratory regulation. Navigenics is just an information service, combining scientifically-published genetic disease correlation data with personal genotype data. [my emphasis]Apparently Navigenics' lawyers think this intriguing argument has a chance of winning over the health department; but just in case that doesn't work, they have a more conventional second line of defense that sounds pretty similar to 23andMe's argument above. Navigenics has been using a CLIA-certified lab since it launched in April and has emphasised its use of a pet physician who rubber-stamps every ordered test, which it feels is sufficient to place it on the right side of Californian regulations. We'll see what the health department makes of these arguments. I can imagine several ways this might all pan out (not being a lawyer, I won't pretend I have much idea how likely any of them are). Firstly, the more respectable, well-backed companies (the big three and Knome) may well be able to hack out a compromise deal with the department and continue operating under only slightly more stringent regulations, while the bottom-dwellers (e.g. the nutrigenetics companies) are forced to move their business to more accommodating regulatory climates. This is probably the best outcome we can hope for - consumers still have the freedom to analyse their own genome without asking their doctor's permission, the department walks away with a sense that the industry has been cleaned up, and the science-free scammers that blight the industry are dealt an important blow. Secondly, insanely, the department could insist that customers need face-to-face consultation with a real live doctor to order any genetic tests. If they stuck to their guns, you could basically kiss goodbye to the fledgling personal genomics industry in California: faced with the obstacle of seeking a doctor's approval to have their scan (rather than five minutes on the web with a credit card) most potential customers simply won't bother. If this legislation spread to other states the industry as a whole would be set back by years. Finally, the department might only push for very strong regulation of any health-related genetic test but leave other forms of genetic testing (ancestry and non-disease traits) alone. The result of this would be ironic: Navigenics, which adopted a serious clinical-centred demeanour from the beginning to steer clear of charges of frivolity that might bring regulatory attention, would be hit much harder than the more relaxed 23andMe. Navigenics has very deliberately avoided doing anything that looked remotely like 23andMe's brand of "recreational genomics" - but that has left them with nothing to fall back on should regulation clamp down hard on health-focused genetics. 23andMe, on the other hand, could probably get by purely on the basis of the non-disease components of their package. Anyway, the next week will be a pretty interesting window in the young life of the personal genomics industry - the decisions made now will likely have major effects on the way the industry evolves over the next few years, in the lead-up to the era of cheap whole-genome sequencing. Stay tuned... |
| A Predictive Tool for Diabetes [Epidemix] Posted: 25 Jun 2008 03:17 PM CDT
The idea behind the Tethys test, called PreDx, is to create a tool that can accurately identify those at increased risk for developing type II diabetes. Diabetes, we know, is one of the fastest growing diseases in the country (and world), accelerated by the upsurge in obesity. 24 million Americans have diabetes, with another 2 million cases diagnosed annually. 60 million Americans are at high risk of developing diabetes, many of these obese or overweight. The traditional tool for diagnosing the disease, as well as for diagnosing a *risk* for developing the disease, is a blood glucose test. The so-called “gold standard” test, a fasting blood sugar value of 140 mg/dl constitutes diabetes, while normal levels run between 70-110 mg/dl. You can see the issue here: What does a value of between 109 and 139 mean? This is the problem with these firm cut-offs - their you-have-it-or-you-don’t nature means that you’re failing to capture people until they have a disease. We’re missing the opportunity to get ahead of illness and maintain health. In the last couple years several genome-wide association studies have identified certain genetic variants with diabetes, to great fanfare. But the problem with these associations is that the rest of the puzzle is, to mix metaphors, blank. We don’t yet know what context these associations exist in, so the seven or eight markers that have been identified may be the complete span of genetic influence, or they may be seven or eight of 1,000 markers out there. In other words, there’s lots of work to do there still. A more traditional attempt at early detection has been the diagnosis of metabolic syndrome, which I’ve written about lots. In a nutshell, metabolic syndrome is an attempt to establish some cutoffs - from glucose, blood pressure, waist circumference - to define a disease that’s a precursor to other disease (namely diabetes and heart disease). It’s an ambitious extrapolation of our ability to quantify certain biological markers, but it’s inexact and, the argument goes, hasn’t proven any better at actually identifying those at risk than blood glucose alone. In other words, it has defined a pre-disease state without actually changing the outcome (at least, that’s the argument; it is a subject of great debate). OK, so that’s the backdrop: a single conventional test that identifies disease better than risks, an emerging but incomplete measure of potential risk, and a measure of pre-disease that has ambiguous impact on the disease. So how about something that identifies risk accurately enough and early enough and strongly enough that it actually impacts the progression towards disease? That’s the idea behind PreDx. The test itself is an ELISA test, which for you microarray junkies may seem disappointing. ELISAs are nothing fancy, they’ve been used for nearly 40 years to detect proteins. The cool part, though, is what goes into that test. Tethys scanned through thousands of potential biomarkers that have been associated with components of diabetes - obesity, metabolic disorder, inflammation, heart disease - and settled on a handful that all closely correlate with diabetes. That’s the ELISA part, testing for levels of those five or so biomarkers. Then second stage of the test is the algorithm: a statistical crunching of the various levels and presence of those markers, to arrive at a Diabetes Risk Score. The DRS is a number between 1 and 10, shown to the tenth of a point, that equals a risk for developing type II diabetes over the next five years. a 7.5 equals a 30% risk of developing diabetes within 5 years, a 9 equals a 60% risk. (the risk for the general population is about 12%, equal to a 5.5 on the PreDx scale) So what’s cool here is the algorithm. Unlike many new diagnostic tests, the smarts aren’t necessarily in the chemistry or the complexity of the technology (it’s not quantum dots or microfluidics or stuff like that). The smarts are in the algorithm, the number crunching. Basically, the test lets the numbers do the work, not the chemistry. At $750 a pop, the PreDx test is too expensive to be used as a general screening test - it’s best used by physicians who’ve already determined their patient is at an increased risk, through conventional means. Tethys says that’ll save $10,000 in healthcare costs on the other end. In other words, it’s a way to pull people out of that pre-diabetes pool, spot their trajectory towards disease, intervene, and avoid onset. In other other words, it’s a tool to change fate. Which is kinda impressive. Another interesting thing here is the simplicity of the 1 to 10 scale. Obviously, this is the work of the algorithm; the actual data doesn’t neatly drop into a 4.5 or a 7.3 figure, it must be converted into those terms. That in/of itself is a complicated bit of biostatistics, and it’s beyond me to assess how they do it. But the fact that an individual will be presented with a Diabetes Risk Score of, say, 8.1, and then shown a chart that very clearly puts this at about a 40% risk of developing diabetes - well, that’s a lot easier for a lay person to make sense of than a blood glucose level of 129 miligrams-per-deciliter. Heck, it’s a lot easier for a *physician* to make sense of. What’s more, this is a quantification of *risk*, not a straight read of a biological level. That’s a very different thing, much closer to what we want to know. We don’t want to know our blood glucose level, we want to know what our blood glucose level says about our health and our risk for disease. The closest physicians can usually get us to *that* number is to go to general population figures - in the case of diabetes, that 12% risk figure for the general population. What PreDx represents, then, is how we are moving from a general risk to a personalized number. This isn’t the abstract application of population studies to an individual, it’s the distillation of *your* markers, using statistical analysis to arrive at an individual risk factor. So I find this pretty compelling. Tethys is developing other predictive diagnostic tests for cardiovascular disease and bone diseases, but far as I know, there are not many similar predictive diagnostic tests out there, for any disease. As mentioned, we have the genomic assocations, which are coming along. Anything else somebody can clue me in on? Lemme know… |
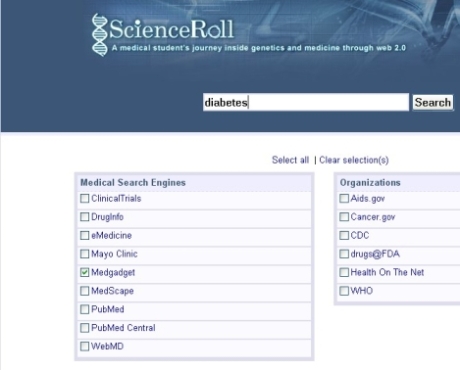
| Personalized Medical Search Engine: With Medgadget [ScienceRoll] Posted: 25 Jun 2008 03:10 PM CDT I’ve already presented a personalized medical search engine, Scienceroll Search, I’m working on with Polymeta.com. You can choose which databases to search in and which to exclude from your list. It works with well-known medical databases and there is an open discussion about which new sources to use. Essentially it’s up to the users. Now you can search in the database through Medgadget posts as well. For example, if you would like to find diabetes-related medical tools and gadgets, just click on Medgadget and make a search: And you not just get some relevant results, but you can use the topic clusters to get closer to the information you need. Let us know if you would like to add a new resource to our list. This is a cross-post with Medgadget. Further reading:
 |
| Posted: 25 Jun 2008 12:55 PM CDT Dear Governor Jindal, As scientists from across the country and members of the Public Affairs Committee of the American Society of Plant Biologists, we are concerned about the passage of bill SB 733 in the Louisiana legislature. In particular, we are concerned that this bill may represent an attempt to introduce religion into science classes. Teachers already try to promote critical thinking skills; thus, why is there a need for a bill to "to create and foster an environment within public elementary and secondary schools that promotes critical thinking skills, logical analysis, and open and objective discussion of scientific theories being studied including, but not limited to, evolution?" Science, as you know as one of the few governors with an undergraduate degree in biology, is based on the interpretation of actual data. Denying the validity of evolutionary theory requires either a complete lack of knowledge of the data and science behind this theory, or purposely ignoring that data and science. It is true that scientists still debate the mechanistic details of the evolutionary process. That is the nature of science: to seek to understand a process in ever greater detail. In the past 150 years, our understanding of the mechanistic details of how evolution occurs has increased tremendously. For example, Darwin did not know the molecular basis of heredity, and it was not until the late 1940's through the early 1950's that DNA was found to be the genetic material. However, the occurrence of scientific debates about the mechanistic details of the evolutionary process does not cast any doubts on the validity of evolutionary theory. In science, a "theory" is not a hunch or a guess, it is an idea that is continually scrutinized with respect to all of the data that scientists accumulate. Over the past 150 years since Charles Darwin published 'On the Origin of Species,' all of the data collected has provided additional support for the validity of the theory of evolution. Indeed, evolution is as clearly and strongly supported by the data as any theory in science. Simply put, there is no controversy whatsoever about the validity of evolutionary theory among honest scientists who are familiar with the data; rather, there is complete agreement that evolution has been the process that has resulted in, to quote Charles Darwin, the "endless forms most beautiful" that live on our planet. As Governor you are in a position of leadership. Your leadership could guide Louisiana down a path where science is taught in science classrooms and scientific issues are discussed within the framework of actual scientific data. We hope that you will not let Louisiana sanction a path designed to mislead students about what science is and how it works. The students of your state deserve the opportunity for a proper science education. Sincerely, Gary Stacey, Chair, University Of Missouri Richard Amasino, University Of Wisconsin Sarah Assmann, Penn State Daniel Bush, Colorado State University Roger Innes, Indiana University James N. Siedow, Duke University Ralph S. Quatrano, Dean of the Faculty of Arts & Sciences, Washington University in St. Louis Thomas D. Sharkey, Chair Dept. of Biochemistry, Michigan State University David E Salt, Purdue University Mary Lou Guerinot, Dartmouth College Roger Hangarter, Indiana University Robert L. Last, Michigan State University Richard T. Sayre, Ohio State University Robert L. Last, Michigan State University Peggy G. Lemaux, University of California, Berkeley Pamela Ronald, University Of California, Davis C. Robertson McClung, Dartmouth College cc Louisiana legislature |
| Finch 3: Getting Information Out of Your Data [FinchTalk] Posted: 25 Jun 2008 12:45 PM CDT |
| Complex disease genes and selection [Yann Klimentidis' Weblog] Posted: 25 Jun 2008 11:24 AM CDT The premise of the paper below is to improve on the use of Dn/Ds (the ratio of nonsynonymous to synonymous mutations) as a measure of purifying selection since it has shown mixed results with respect to explaining differences between disease related genes and other genes. They argue that this may be a result of an outdated OMIM database. Here they "hand curate" their own version of the OMIM, using only well defined highly penetrant diseases. for simple disease associated genes: Dn/Ds values tend to be higher in genes with recessive disease mutations (median= 0.184, n = 452) than in those with dominant disease mutations (median = 0.084, n = 294)for genes associated with complex disease: genes associated with complex-disease susceptibility tend to be under less-pervasive purifying selection than are other classes of essential or disease genes.They argue that: ... existing data raise the possibility that, whereas simple disorders are generally well-described by models of purifying selection, complex-disease susceptibility is tied, at least in part, to evolutionary adaptations.Update: I just noticed that Razib also has a discussion of this paper Natural Selection on Genes that Underlie Human Disease Susceptibility Ran Blekhman, Orna Man, Leslie Herrmann, Adam R. Boyko, Amit Indap, Carolin Kosiol, Carlos D. Bustamante, Kosuke M. Teshima, and Molly Przeworski Current Biology Vol 18, 883-889, 24 June 2008 Abstract: What evolutionary forces shape genes that contribute to the risk of human disease? Do similar selective pressures act on alleles that underlie simple versus complex disorders [1, 2, 3]? Answers to these questions will shed light onto the origin of human disorders (e.g., [4]) and help to predict the population frequencies of alleles that contribute to disease risk, with important implications for the efficient design of mapping studies [5, 6, 7]. As a first step toward addressing these questions, we created a hand-curated version of the Mendelian Inheritance in Man database (OMIM). We then examined selective pressures on Mendelian-disease genes, genes that contribute to complex-disease risk, and genes known to be essential in mouse by analyzing patterns of human polymorphism and of divergence between human and rhesus macaque. We found that Mendelian-disease genes appear to be under widespread purifying selection, especially when the disease mutations are dominant (rather than recessive). In contrast, the class of genes that influence complex-disease risk shows little signs of evolutionary conservation, possibly because this category includes targets of both purifying and positive selection. |
| Where You Get Your Mammogram Matters! [Cancer and Your Genes] Posted: 25 Jun 2008 10:17 AM CDT A research group led by Dr. Stephen Taplin of the National Cancer Institute has published a paper in the June 18th issue of the Journal of the National Cancer Institute that assesses whether certain factors differing between mammography facilities are associated with better interpretive accuracy on screening mammograms. The bottom line is that there are several things that were signficant. Sensitivity, the ability to detect breast cancers, was quite high and varied little. However, specificity and positive predictive value varied significantly between sites. Thus, it seems that the degree to which sites may be "overcalling" their reads (presumably to avoid missing something) varies. The following mammography facility traits were associated with better interpretive accuracy for screening mammography:
The authors note that it was surprising that double reading did not improve overall test accuracy and that this contrasts with the results of randomized clinical trials. It may be that double reading methods differ or have been implemented differently in typical clinical practice environments outside of clinical trials. This deserves a closer look in the future. Given the relatively weak effect of this predictor of performance, the authors suggested that it should not be utilized to differentiate amongst facility performance until further detailed research is done to sort out the controversy. It is important to keep in mind that the above factors affected specificity and positive-predictive value primarily. The authors point out that whether sensitivity or specificity is most important in picking a mammogram facility is dependent on a value judgment. That is, some women may be most concerned about sensitivity at all costs (i.e., a higher false positive rate leading to a biopsy does not matter). Other women may wish to consider factors like those noted above that affect specificity and positive predictive value (i.e., they would like to avoid unnecessary biopsies while keeping sensitivity high). The results of this retrospective study should be confirmed elsewhere before they are utilized for decision-making. Nevertheless, you may want to have a dialogue with your physician about mammography facility choice. |
| Tangled Bank #108. [T Ryan Gregory's column] Posted: 25 Jun 2008 09:04 AM CDT The current edition of Tangled Bank, the blog carnival about evolution, is now up at Wheat-dogg's World. |
| 454 anew......It's been 1 year [The Gene Sherpa: Personalized Medicine and You] Posted: 25 Jun 2008 07:04 AM CDT |
| Bird Flu Flap [Sciencebase Science Blog] Posted: 25 Jun 2008 07:00 AM CDT
There are just so many different types of host within which novel microbial organisms and parasites might be lurking, just waiting for humans to impinge on their marginal domains, to chop down that last tree, to hunt their predators to extinction, and to wreak all-round environmental habitat on their ecosystems, that it is actually only a matter of time before something far worse than avian influenza crawls out from under the metaphorical rock. In the meantime, there is plenty to worry about on the bird flu front, but perhaps nothing for us to get into too much of a flap over, just yet. According to a report on Australia’s ABC news, researchers have found that the infamous H5N1 strain of bird flu (which is deadly to birds) can mix with the common-or-garden human influenza virus. The news report tells us worryingly that, “A mutated virus combining human flu and bird flu is the nightmare strain which scientists fear could create a worldwide pandemic.” Of course, the scientists have not discovered this mutant strain in the wild, they have simply demonstrated that it can happen in the proverbial Petri dish. Meanwhile, bootiful UK turkey company - Bernard Matthews Foods - has called for an early warning system for impending invasions of avian influenza. A feature in Farmers Weekly Interactive says the company is urging the government and poultry industry to work together to establish an early warning system for migratory birds that may carry H5N1 avian flu. “Armed with this knowledge, free range turkey producers would be able to take measures to avoid contact between wild birds and poultry.” That’s all well and good, but what if a mutant strain really does emerge that also happens to be carried by wild (and domesticated birds) or, more scarily by another species altogether? Then, no amount of H5N1 monitoring is going to protect those roaming turkeys. While all this is going on, the Washington Post reports that the Hong Kong authorities announced Wednesday (June 10) that they are going to cull poultry in the territory’s retail markets because of fears of a dangerous bird flu outbreak. H5N1 virus was detected in chickens being sold from a stall in the Kowloon area and 2700 birds were slaughtered there to prevent its spread. In closely related news, the International Herald Tribune has reported that there has been an outbreak of bird flu in North Korea. “Bird flu has broken out near a North Korean military base in the first reported case of the disease in the country since 2005, a South Korean aid group said Wednesday.” But, note, “since 2005″, which means it happened before, and we didn’t then see the rapid emergence of the killer strain the media scaremongers are almost choking to see. Finally, the ever-intriguing Arkansas Democrat Gazette reported, with the rather uninspiring headline: Test shows bird flu in hens. Apparently, a sample from a hen flock destroyed near West Fork, Arkansas, tested positive for avian influenza. A little lower down the page we learn that the strain involved is the far less worrisome H7N3. So, avian influenza is yet to crack the US big time. Thankfully. A post from David Bradley Science Writer |
| What does DNA mean to you? #11 [Eye on DNA] Posted: 25 Jun 2008 03:09 AM CDT
|
| TGG Interview Series VI - Ann Turner [The Genetic Genealogist] Posted: 25 Jun 2008 02:00 AM CDT Ann Turner has been a member of the genetic genealogy community since 2000, and during that time she has made great contributions to field (as will become obvious from her interview). According to her brief biography at the Journal of Genetic Genealogy:
As stated in her bio, Ann is the co-author of “Trace Your Roots With DNA", the premiere book on genetic genealogy (the other co-author, Megan Smolenyak Smolenyak, was featured earlier in this series). Ann continues to contribute frequently to the GENEALOGY-DNA mailing list at Rootsweb, and has been especially active in genetic genealogical analysis of new SNP testing by companies such as 23andMe and deCODEme. Once again, I highly recommend subscribing to the GENEALOGY-DNA mailing list if you are interested in genetic genealogy testing! In the following interview, Ann discusses her introduction to genetic genealogy, some of her experiences with testing, and the use of large-scale SNP testing for genealogical purposes. TGG: How long have you been actively involved in genetic genealogy, and how did you become interested in the field? Ann Turner: I’ve been actively involved since the year 2000, when DNA testing for the ordinary consumer first came to market. I had been waiting for that moment for a long time, though. I was first inspired by an article in the NEHGS magazine by Thomas Roderick, Mary-Claire King, and Robert Charles Anderson. It was the first to point out the potential of tracing long matrilineal lines with mtDNA. That was written clear back in 1992, so it took a while for my dream to become reality. I wanted to have someone to chat with about this new field, so I founded the GENEALOGY-DNA mailing list. Be careful what you wish for! The list now carries thousands of messages per month. But it was also the means by which I “met” Megan Smolenyak, my co-author for “Trace Your Roots with DNA” and countless other wonderful fellow travelers in this strange new land. TGG: Have you undergone genetic genealogy testing? Were you surprised with the results? Did the results help you break through any of your brick walls or solve a family mystery? AT: Yes, I’ve experimented with many different types of tests. One of the most satisfying endeavors was learning the real surnname and origins of a great-grandfather, who was orphaned at a young age. There were family legends that he had a half-brother, who was taken in by another family and never heard from again. Through traditional genealogy research, I tracked down a potential descendant and ordered a Y-DNA test for him and a cousin of mine. The result was a perfect match. The next step was to connect this family to a Lancaster County, Pennsylvania line, which traced its origins and an unusual spelling of the surname back to 1740. I simply put out a call for any male named Shreiner, and the respondent was also a perfect match. Again, this technique was combined with traditional genealogical research, which always goes hand-in-hand with DNA testing, but it was the DNA that enabled me to span centuries: 150 years forward to a descendant of the half-brother, and 150 years back to the origins of the surname in the United States. TGG: I know that you have been analyzing the results of large-scale genome scanning tests by 23andMe and deCODEme, and I was wondering what your thoughts are regarding the applicability of these results to genetic genealogy. Will these SNP tests shed light on the human Y-DNA or mtDNA trees, or should we just wait a few years for full-genome sequencing? AT: The mtDNA and Y-SNP tests from the genome scans are no substitute for the mtDNA and Y tests offered through the genealogically oriented companies, which offer much greater resolution. I regard those features as fringe benefits of the scans, which provide access to an unprecedented amount of autosomal data. Someday it may be possible to trace small segments of autosomal DNA (”haplotype blocks”) to a common ancestor. That will require massive databases and massive computational power! Sorenson Molecular Genealogy Foundation is pioneering in this new domain. TGG: Thank you Ann, for a terrific interview!
|
| Pacemaker: The Program that puts the heart back into the Health Care Industry [Think Gene] Posted: 25 Jun 2008 01:48 AM CDT Warning: explicit language Source: Grand Theft Auto 4: Public Liberty Radio It’s funny, it’s crude, but it’s interesting: When joining an HMO to get health care is mandatory, when the forces-that-be have every legal advantage, when the fine-print is written intentionally to be incomprehensible, what checks HMO’s incentive to bill premiums yet not pay for care? When all of health and biochemistry is too complicated for even doctors (hence, specialization), when advertising has evolved into audiovisual crack, when the first obligation of a drug company’s officers is to its shareholders, what checks drug companies from producing addictive consumerist luxuries in the pursuit of greatest profit? And when everyone credible has so must vested in the status quo, the only dissent is crazy. Quotes!Wilson Taylor Sr. of Copay Health Systems (HMO):
Sheila Stafford of Betta Pharmaceuticals (drug company):
Waylon Mason, The Down-Home Medicine man and Homeopathic (alternative medicine):
(is this a better podcast that Helix Heath’s?) |
| Snake Oil Alert: Blue Light Cancer Treatment at ThinkGene [Cancer and Your Genes] Posted: 25 Jun 2008 01:33 AM CDT Here's a post from Josh Hill at ThinkGene about an unpublished study of Blue Light effect on tumors in mice. It's a hands-down winner of today's Snake Oil of the Day Award. Lest we give cancer patients and the general public false hope and the wrong impression, it might be wise to let one like this at least get through peer review before sensationalizing it... |
| Riding the open wave… [My Biotech Life] Posted: 24 Jun 2008 11:44 PM CDT
Aside from getting to know the great people responsible for creating OpenWetWare, I have had the opportunity to directly participate in this great project at a core level with the implementation of specific tools like the lab notebook and interact with the great community that makes OWW such an incredible online resource. I have recently taken on the responsibility of spearheading the OWW Community blog where I’ll be publishing among many things, projects that take place on OWW, tips and tricks that can be used on the wiki, interesting news from the Open Science community and much more. If you are interested in topics such as experimental biological research, sharing of scientific knowledge, web tools for collaborative research and other similar subjects, I recommend you subscribe to the OWW Community blog. I hope to see you there. Also, if you happen to be a twitter user, feel free to follow my and/or OpenWetWare’s tweets! Last but not least, if you aren’t part of the OpenWetWare community yet, what are you waiting for? Post from: My Biotech Life |
| Blue light used to harden tooth fillings stunts tumor growth [Think Gene] Posted: 24 Jun 2008 10:50 PM CDT |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |




 I’m not entirely convinced that bird flu (avian influenza) is going to be the next big emergent disease that will wipe out thousands, if not millions, of people across the globe. SARS, after all, had nothing to do with avians, nor does HIV, and certainly not malaria, tuberculosis, MRSA, Escherichia coli O157, or any of dozens of virulent strains of disease that have and are killing millions of people.
I’m not entirely convinced that bird flu (avian influenza) is going to be the next big emergent disease that will wipe out thousands, if not millions, of people across the globe. SARS, after all, had nothing to do with avians, nor does HIV, and certainly not malaria, tuberculosis, MRSA, Escherichia coli O157, or any of dozens of virulent strains of disease that have and are killing millions of people.


No comments:
Post a Comment