The DNA Network |
| What’s on the web? (18 August 2008) [ScienceRoll] Posted: 18 Aug 2008 04:11 PM CDT The conference schedule of the Medicine 2.0: Social Networking and Web 2.0 Applications in Medicine and Health is up. The medical bloggers’ panel and my oral presentation both will take place on the first day.
 |
| Web 2.0 and Medicine Course: First Part [ScienceRoll] Posted: 18 Aug 2008 03:42 PM CDT As you may remember, I will run the first medicine 2.0 course from this September at the University of Debrecen. Now I’m asking you to add your ideas and suggestions regarding the subjects. This time, we should focus on the first two weeks: 1st week:
2nd week:
If you have anything in mind, any websites or services I should talk about in my presentations, please let me know. I will feature this experiment in the first slideshow as an example about how web 2.0 works.  |
| Power-up Your Restriction Screens [Bitesize Bio] Posted: 18 Aug 2008 12:18 PM CDT At the end of the day, all you want to know from a restriction screen is whether your insert is in the vector. But while the standard "chop out a fragment" approach favored by most researchers provides good information about the presence of the insert and its orientation, it only uses a portion of the potential power of a restriction screen. It doesn't take much more work to set up a restriction screen that is more informative, allowing you to confirm a negative result and the identity of your vector, at the same time as checking for your insert. Here's a few ways you can soup-up your restriction digest. 2. To test positive results: If you want a quick test of whether your insert has actually been cloned in, a good approach is to chop out a fragment from the insert itself. This way you can simultaneously verify the presence of an insert and its identity. This can be especially useful if the cloning-in restriction sites have been damaged during cloning. Let us know your restriction digest tips in the comments. |
| MUST LISTEN: Robert Krulwich on Science for the Masses [The Daily Transcript] Posted: 18 Aug 2008 10:31 AM CDT If you are a scientist, I urge you to listen to the commencement speech that Krulwich gave to the Cal Tech graduates earlier this year. Read the comments on this post... |
| Polarity, Diffusion, and Cellular Aging [Bitesize Bio] Posted: 18 Aug 2008 08:56 AM CDT Two recent articles that I came across clearly illustrate ways in which cellular asymmetry is both easily established by basic factors, and provide the basis for processes like cellular polarity and aging. One cannot claim with certainty what these findings in mathematical models and yeast, respectively, impart to our understanding of human health. But they do allow us to generally describe very basic rules for the operation of eukaryotic life. In the second article, Zhanna Shcheprova and coworkers in Zurich, Switzerland, illustrate a mechanism for asymmetric segregation of age during yeast budding. They show that a diffusion barrier develops in the nuclear envelope of the dividing yeast nucleus. The barrier prevents pre-existing nuclear pores and other membrane-associated proteins from moving into the bud. This second article provides an excellent example of the spontaneous emergence of asymmetry/polarity, and an example of its function (i.e. cell aging). In this case though, cytoskeleton-based transport is replaced with compartmentalization - really a variation on transport. And that’s really what the aging article shows - a variation on a biological “rule” that’s been uncovered. It could have turned out any number of ways, and now that it seems so patently obvious, it almost seems like a tautology. But it’s not. Polarity didn’t have to be generated by diffusion. At the level of our basic building blocks, complex life changes such as aging, locamotion, and others, are becoming rather, well, simple. Image: Figure 1 from Altschuler et al., Nature
|
| Red-hot Alchemist [Sciencebase Science Blog] Posted: 18 Aug 2008 07:00 AM CDT
Van Gogh was two-timing his canvas, the Alchemist learns this week, thanks to novel X-ray studies of a seemingly innocuous piece called Patch of Grass, which hides a woman’s face beneath its green and peasant landscape. Professional wine tasters and vintners with a penchant for pepping up their plonk should have something new to worry about thanks to the development of an electronic tongue for detecting adulterated wines and those labeled with the wrong vintage. In biochemistry, sex and sleep turn out to be inextricably entangled, at least in the world of the lab technician’s favorite nematode worm, Caenorhabditis elegans. Traditional Chinese Medicine is heavily marketed despite a lack of clinical evidence of efficacy of many of the remedies. However, The Alchemist hears of a traditional remedy for allergy that, toxic components removed, could work to prevent life-threatening peanut allergy. The world of red hot chili peppers wouldn’t be so hot if it were not for nibbling insects and a fungus that infects the chilis. Finally, a million-dollar grant to get the blood pumping will for the next five years fund research into how the brain controls blood pressure and could eventually lead to new treatments for hypertension and cut deaths from cardiovascular disease. a |
| The New BRCA....this time its the Colon!!! [The Gene Sherpa: Personalized Medicine and You] Posted: 18 Aug 2008 05:53 AM CDT |
| SciFoo: scientific fireworks [The Seven Stones] Posted: 18 Aug 2008 04:37 AM CDT
SciFoo is a so-called 'unconference': there is no program or more precisely, as Timo Hannay explained during the opening of the conference, the attendees are the 'program'. The actual schedule was defined only on the first evening in a purposefully chaotic process by anyone who wished to organize a session on any topic. For the next two days, in a festival of parallel sessions, astrophysicists, 'googlers', technologists, molecular biologists, taxonomists, game designers, flying car constructors, publishers, thinkers and (some) dreamers discussed and exchanged ideas with great enthusiasm and a rare intensity and openness. Needless to say that deciding which session to attend was close to impossible... In any case, I ended up following three types of talks: a series on systems biology related topic (data integration, machine learning, personal genomics, baroque structure of the transcribed genome), several (of many) sessions focused on the theme of open data/science and finally some more eclectic sessions (only from my standpoint, of course) on diverse topics such as the foundations of the concept of time in physics, on some demonstration of very simple yet powerful Python scripting exercises to analyze text and the potential of game design to harness our 'cognitive surplus'. I cannot possibly summarize all the talks, interactions and impressions gathered at this meeting, but here are a few subjective excerpts:
The meeting ended with some final scientific fireworks, when some of the speakers gave a series of brilliant 2 min summary talks, providing a colorful overview of the many sessions we inevitably had missed. I have to admit that I like fireworks and I would certainly have enjoyed having a little more of this final kaleidoscopic view of science. Clearly, the authentic value of this conference lies in the unique and direct human interactions, but I wish there would be nevertheless some way–perhaps by using this last session in some form of outreach action–to disseminate this pure joy of scientific diversity and curiosity to a broader audience. Credits: illustrations from Bob Lee, Flickr, some rights reserved |
| "Free the Gene!" Fluorescent Black Graphic Novel Interview [Genome Blog] Posted: 18 Aug 2008 03:25 AM CDT Here’s an interesting Biopunk find! I was scanning the shelves at the local gas station and came across the September issue of Heavy Metal Magazine. As I flipped through the pages, I came upon a graphic novel intro called Fluorescent Black by Author Matt F. Wilson. As fate would have it, I was honored to meet Matt through the wonders of the internet and myspace. He graciously agreed to do an interview for this Blog and the interview in full is posted in the read more link below.
|
| To fix or not to fix, that is the question [Mailund on the Internet] Posted: 18 Aug 2008 03:23 AM CDT Ok, I tried my fix to the BiRC webpages, but I am not sure I should have. After the server process was restarted, all the auto-generated files were gone, and now I don’t know how to get them back. The good news is that loading the pages will no longer crash your browser. The gigantic files are the ones that are completely missing now. The pages without the stylesheets look like crap, though. |
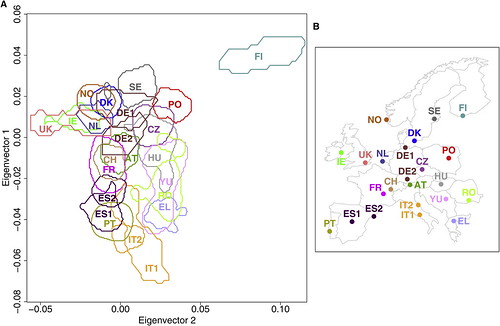
| How well does your genome predict your postcode? [Genetic Future] Posted: 18 Aug 2008 03:19 AM CDT Well, it's far from GPS precision, but the concordance between this genetic map of Europe (below left) and the physical sampling locations of populations throughout Europe (below right) is pretty good for a first draft:
Dienekes has an excellent discussion of the technical details, while Razib has labelled a plot showing all of the individuals in the study to make it easier to assess the degree of scatter and overlap. The take-home message: rather than being one homogeneous mass, Europeans in fact show considerable population substructure, such that genetic information can be used to roughly predict geographical ancestry. An analysis of just a few hundred thousand genetic markers (i.e. less than is currently offered by personal genomics companies 23andMe or deCODEme) would be more than adequate in most cases to distinguish a Pole from a Parisian, or a Swede from a Spaniard. (To be more precise, it would be sufficient to discriminate between individuals for whom most ancestors were natives of these regions; recent migrants will obviously be misclassified.) What drove these genetic differences? Mostly it will have been chance - random increases or decreases in the frequency of markers throughout the genome accumulated over a few millennia of genetic isolation. But at least some of these differences have been driven by natural selection: for instance, the lactase gene LCT, which has been subject to strong selection to allow lactose digestion in adults in populations reliant on dairy agriculture, represents 9 out of the top 20 most differentiated markers; a marker in the gene HERC2, which is associated with eye colour variation and has been under selection in Europeans and Asians, comes in at number 19. This indicates that at least some of the genetic - and thus physical and possibly behavioural - differences between the various European populations stem from evolutionary adaptation to their local environments. I'll leave the technical commentary to Dienekes, but I do want to make one important point: the accuracy of the map will have been limited by the fact that the markers used in this study represent sites of common variation; data from large-scale genome sequencing will generate far, far better maps. The major reason for this is that sequencing will provide information on rare, highly spatially-restricted variants - many of which will be limited to single families and thus be extremely informative about geographical ancestry. Basically, if you had complete genome sequences from enough Europeans you could reconstruct the genetic map of Europe with exquisite precision. In addition to empowering genetic genealogists, researchers could use deviations between the genetic and physical maps to make powerful inferences about historical migration events and recent episodes of natural selection. With any luck, this is the sort of data that will simply fall out from large-scale population genomic studies being conducted over the next decade or so. Update: Kambiz at Anthropology.net puts these results in a broader scientific context. Lao et al. (2008). Correlation between Genetic and Geographic Structure in Europe. Current Biology DOI: 10.1016/j.cub.2008.07.049 Image source: Figure 1 from Lao et al. |
| Evolution as the Recycler of the Cell's Tools [adaptivecomplexity's column] Posted: 17 Aug 2008 10:11 PM CDT Part 2 on The Plausibility of Life How does evolution shape living things? The fact that evolutionary forces, such as natural selection, can shape living creatures is well-established, but how malleable those creatures are, and what the increments of change are is less well established. We have a fairly good idea of how genes can change, but how does that genetic change translate into physical changes in the shape and functioning of the organism itself - that is, how does genetic change translate into changes in the organism's phenotype? The authors of The Plausibility of Life, Marc Kirschner and John Gerhart, argue that this issue has been ignored in evolutionary theory (although they go on to say that it was justifiably ignored for a long time - before modern molecular and cell biology, there was no way to effectively address this question):
These questions get to the heart of the evolution of complexity. |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |


 In my
In my  In his list of eight 'generative' values (
In his list of eight 'generative' values (
No comments:
Post a Comment