The DNA Network |
| Christina and Jessica beat BRCA1 [The Gene Sherpa: Personalized Medicine and You] Posted: 19 Aug 2008 07:56 PM CDT |
| Complexity of Breast Cancer [www.cancer-genetics.com] Posted: 19 Aug 2008 04:31 PM CDT
Hereditary breast cancer (HBC) accounts for as much as 10% of the total BC burden. Only about 30 percent of these cases will be found to be due to a germline mutations in well known BRCA1 and/or BRCA2 genes, but the rest won’t have these mutations. Less than 10 percent of remaining HBC will fall into other rare conditions - and here we can see breast cancer as heterogeneous disease (ref.): Cowden, Li-Fraumeni syndromes, heterozygosity for Ataxia telangiectasia-mutated gene (ATM) or for CHEK2 1100delT or other rare conditions Nijmegen breakage syndrome (NBS1), familial diffuse gastric cancer (CDH1), Peutz-Jeghers syndrome (STK11), Fanconi anemia (BRIP1, PALB2), Bloom syndrome (BLM)) contribute sligthly - there is consensus for ten most important genes involved in HBC (ref.) An estimated additional 15–20% of those affected with BC will have one or more first- and ⁄ or second-degree relatives with BC (familial or polygenic breast cancer). Therefore, when these numbers are combined, familial BC risk accounts for approximately 20–25% of the total BC burden (see figure). Here we’re talking about so called low-penetrance susceptibility genes and variants (SNPs), like rs2981582 in FGFR2, rs889312 in MAP3K1, rs3803662 in TNRC9, rs1801270 in CASP8 and many more, most of which were hot topics in the recent years. Particular alleles (particular “letter” variants of these digitalized “rs” SNPs) only increase risk slightly (twice or so) and are intense study object now, but they sooner or later will enter clinical practice.  |
| Banned Performace Enhancing Drug Classes [Bayblab] Posted: 19 Aug 2008 03:25 PM CDT  The IOC has banned athletes from using drugs to enhance their performance. What is the purpose of this policy? Surely technology offers athletes many ways to enhance their performance. If the issue is fairness, I don't really see the difference between the new speedo swim suits and performance enhancing drugs. If the issue is athlete health, which seems more reasonable to me, I wonder what the difference is between taking carefully administered drugs and the health of young female gymnasts. From the World Anti-Doping Agency (WADA): "...Clean Athletes are the real heroes who make brave choices everyday by not doping. They deserve competition that is safe and fair. They deserve a level playing field." In any case, back to science, here's a Ben Johnson's breakfast of all that you can't indulge in if your are an elite olympic athlete: Anabolic Agents: Most importantly this includes small molecule steroids related to testosterone. These increase your muscle mass and red blood cells. Great for strength requiring events. Obviously they have masculizing effects aswell. The scary side-effect of shrinking testicles is temporary, however the long term effects are heart related and are probably undesirable. They can also make you grow a beard if your are a woman and initiate male pattern baldness early. This catagory also includes steroids used to treat asthma, Beta-2 agonists. These drugs increase lean muscle mass, and also have some stimulant properties. A therapeutic use exemption can be obtained if you are asthmatic. Blood doping: Increasing the oxygen carrying capacity of your blood by increasing the number of red blood cells (RBCs) in your body. This is great for endurance sports. This can be done by harvesting RBCs, storing them, then readministering to the athlete before competition. This is pretty old school now and more common is the use of erythropoeitin (EPO). EPO is a peptide hormone that stimulates growth of RBCs. It has medicinal uses and therefore is cloned and available. Detection is difficult but possible. Too much RBCs makes your heart work hard and apparently if you are an elite athlete with a low resting heart rate you can die in your sleep. Peptide hormones: There are many hormones that may give an athlete an unfair advantage. The previously mentioned EPO and additionally other growth hormones that increase muscle weight. This includes insulin, and suprisingly I didn't see a therapeutic use exception for exogenous insulin for diabetics. I assume there is. Stimulants: Caffine is fine, but go easy on the cocaine. These increase heart rate and thereby bloodflow and increase metal alertness. I would think the most commonly used as a performance enhancer is ephedrine, mostly because I have heard about it the most. Diuretics: Used to increase urination to loose weight. Good for being the largest guy in the smallest weight catagory. Also used to eliminate drugs from your system, ie a masking agent. Narcotic Analgesics: No pain, no gain. But without the pain it is all gain. Opiates are probably the most commonly used to overcome injury or train for longer periods that would normally cause pain. Gene Doping: This is the newest catagory and I don't really think there are any examples. But my guess is that you could use some techniques to genetically increase EPO or another of the above protein products in an athlete. From WADA: "The non-therapeutic use of cells, genes, genetic elements, or of the modulation of gene expression, having the capacity to enhance athletic performance, is prohibited." If you are wondering about your particular prescription make sure to check the complete list of banned substances from the World Anti-Doping Agency. If it's on the list, increase your dose before any competitive event, winning is everything. |
| Compare and Compare Alike [Sciencebase Science Blog] Posted: 19 Aug 2008 02:30 PM CDT Back in June 2001, I reviewed an intriguing site that allows you to compare “stuff”. At the time, the review focused on how the site could be used to find out in how many research papers archived by PubMed two words or phrases coincided. I spent hours entering various terms hoping to turn up some revelationary insights about the nature of biomedical research, but to no avail. I assumed the site would have become a WWW cobweb by now, but no! compare-stuff is alive and kicking and has just been relaunched with a much funkier interface and a whole new attitude. And as of fairly recently, the site now has a great blog associated with it in which site creator Bob compares some bizarre stuff such as pollution levels versus torture and human rights abuses in various capital cities. Check out the correlation that emerges when these various parameters are locked on to the current Olympic city. It makes for very interesting reading. Since the dawn of the search engine age people have been playing around with the page total data they return. Comparing the totals for “Company X sucks” and “Company Y sucks”, for example, is an obvious thing to try. Two surviving examples of websites which make this easy for you are SpellWeb and Google Fight, in case you missed them the first time around. compare-stuff took this a stage further with a highly effective enhancement: normalisation. This means that a comparison of “Goliath Inc” with “David and Associates” is not biased in favour of David or Goliath. Compare-stuff with its new, cleaner interface now takes this normalisation factor to the logical extreme and allows you to carry out a trend analysis and so follow the relative importance of any word or phrase. For example, “washed my hair”, with respect to a series of related words or phrases, for example “Monday”, “Tuesday”, “Wednesday”…”Sunday”. The site retrieves all the search totals (via Yahoo’s web services), does the calculations and presents you with a pretty graph of the result (the example below also includes “washed my car” for comparison). Both peak at the weekend but hair washing’s peak is broader and includes Friday, as you might expect. It’s a bit like doing some expensive market research for free, and the cool thing is that you can follow the trends of things that might be difficult to ask in an official survey, for example: You can analyse trends on other timescales (months, years, time of day, public holidays), or across selected non-time concepts (countries, cities, actors). Here are a few more examples: Which day of the week do people tidy their desk/garage? At what age are men most likely to get promoted/fired? Which popular holiday island is best for yoga or line dancing?: Which 2008 US presidential candidate is most confident? Which day is best for Science and Nature? As you can see, compare-stuff provides some fascinating sociological insights into how the world works. It’s not perfect though. Its creator, Bob MacCallum, is at pains to point out that it can easily produce unexpected results. The algorithm doesn’t know when words have multiple meanings or when their meaning depends on context. A trivial example would be comparing the trends of “ruby” and “diamond” vs. day of the week. The result shows a big peak for “ruby” on “Tuesday”, not because people like to wear, buy or write about rubies on Tuesday, but because of the numerous references to the song “Ruby Tuesday” of course. However, since accurate computer algorithms for natural language processing are still a long way off, MacCallum feels that a crude approach like this is better than nothing, particularly when used with caution. Help is at hand though, the pink and purple links below the plot take you to the web search results, where you can check that your search terms are found in the desired context; in the top 10 or 20 hits that is. On the whole it does seem to work, and promises to be an interesting, fast and cheap preliminary research tool for a wide range of interest areas. With summer well under way, Independence Day well passed, and thoughts of Thanksgiving and Christmas coming to the fore already (at least in US shops), I did a comparison on the site of E coli versus salmonella for various US holidays. You can view the results live here, as well as tweaking the parameters to compare your own terms. Originally posted June 4, 2007, updated August 19, 2008 a |
| Remember, kids, those vintage SNP chips may be worth something someday… [genomeboy.com] Posted: 19 Aug 2008 11:27 AM CDT Remember cycle sequencing? No? Remember grunge? Jeez Louise, you’ve got to wake up pretty early to beat these folks to the punch. Anyway, as they point out, the September/October issue of Technology Review features an opinion piece by yours truly:
|
| Calling all enthusiasts: Misha Angrist talks personal genomes [The Personal Genome] Posted: 19 Aug 2008 10:46 AM CDT Misha Angrist takes stock of a number of issues related to personal genomics in an article published today. Do people want access to their genomic data? Should people have access? What should they expect to discover from a genome sequence? What has been his experience thus far? Misha also draws attention to one under appreciated aspect of obtaining a personal DNA sequence: there’s more to genomics than personalized medicine. Genomics might also be interesting and worthwhile even without obvious direct benefits.
Sober-faced enthusiasts may be the bootstraps necessary for the field of personal genomics to achieve lift-off. According to Misha:
If amateur enthusiasts can make contributions to nuclear fusion (see video), why not personal genomics? Read the article. – Misha Angrist. Personal Genomics: Access Denied? Even if we can’t interpret the data, consumers have a right to their genomes. MIT Tech Review. September/October 2008 See, Misha’s blog GenomeBoy.com See, PGP-10 profiles |
| The challenges of psychiatric genetics [Genetic Future] Posted: 19 Aug 2008 09:40 AM CDT  Back in April I posted on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year. Back in April I posted on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year.The major message from that article is that although bipolar disorder is massively influenced by genetic factors (around 85% of the variation in risk is thought to be due to genetics) we still don't really have the faintest idea exactly which genes are involved. This is despite three reasonably large genome-wide association studies involving over 4,000 bipolar patients in total, which generated weak and contradictory results and failed to provide a single compelling candidate for genetic variation underlying this disease. This disappointing result has also held largely true for other psychiatric conditions with strong genetic components, such as schizophrenia, major depression and autism. Genetic studies of these conditions have had some success identifying rare mutations that underlie severe cases, but the vast majority of the genetic variants contributing to risk remain undiscovered. There are several reasons why genome-wide association studies can fail to yield significant harvests of disease-associated genes. I summed these up with respect to bipolar disease as follows: The researchers are surely hoping that small effect sizes are the major problem, since this is the easiest problem to remedy (simply increase sample sizes). Disease heterogeneity - in other words, multiple diseases with distinct causes that all converge on a bipolar end-point - also seems like a particularly plausible explanation given the complexities of mental illness. It's also likely that various types of genetic variants that are largely invisible to existing SNP chips, like rare variants and copy-number variation, are important.The same story probably holds largely true for other psychiatric conditions. In this week's issue of Nature, a news article and an editorial both tackle the challenges of psychiatric genetics, and lay out the ambitious strategies currently being pursued by researchers around the world to overcome them. Small effect sizes The first hurdle that I describe above is the fact that most of the variants underlying these conditions probably have very small effect sizes (only increasing risk by less than 20%). Such variants will only be identified by cranking up sample sizes immensely, an approach that has yielded some limited success for other genetically complex traits such as height and obesity. The Nature news feature has a table listing some of the major collaborative efforts currently collecting genetic information from the very large cohorts required to dissect out the basis of these conditions: 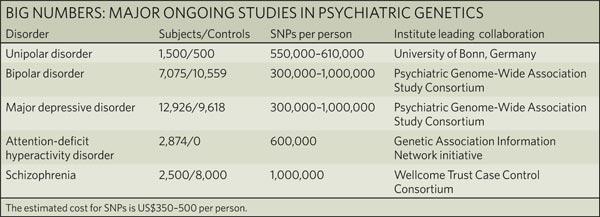 In most cases, these samples are being built up by pooling results from multiple different studies, often gathered by groups from around the world. As sample sizes increase the power of studies to detect small-effect variants grows. The effect of sample size on the power of genome-wide association studies is illustrated in the graph below from a recent review by Peter Visscher*: 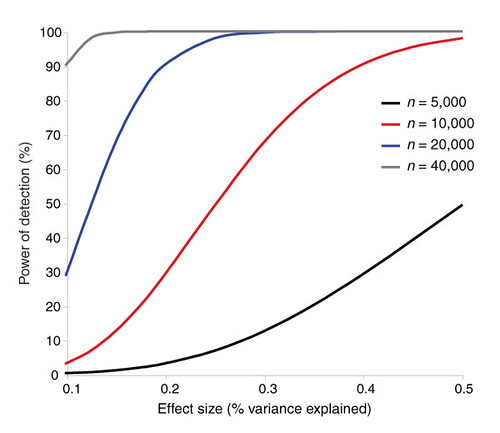 Take a single genetic variant that explains just 0.5% of the variance in the risk of a psychiatric disorder. With a sample size of 5,000 individuals with that disorder you still have a mere 50% chance of detecting that variant. Double your sample size, and that probability jumps to a near-certainty of detection - and your power of detecting even smaller-effect variants (explaining, say, 0.2% of the risk) starts to climb to respectable levels. By staring at those curves for a while, and bearing in mind that many of the variants found by recent genome-wide association studies explain well under 0.2% of the risk variance, you will quickly start to appreciate why researchers are pushing for ever-larger disease populations to work with. With truly enormous samples on the order of 50 to 100 thousand patients - not out of the question for international consortiums studying reasonably common diseases such as bipolar - the power to detect even very weak risk variants becomes reasonable. If there are common genetic variants contributing to the risk of these diseases, such large collaborative studies will eventually find them; so long, of course, as they can tackle the next (and potentially far more serious) problem of disease heterogeneity. Complex, heterogeneous diseases  The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible. The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible.This complexity and heterogeneity is the basis of considerable tension between geneticists and neuroscientists, which is explored in the Nature editorial. Basically, to build up those massive sample sizes shown above geneticists are forced to lump together patients with a variety of clinical symptoms, thus essentially ignoring the complexity inherent in these conditions - a failure that neuroscientists find inexcusable. In turn, geneticists (like myself) get seriously annoyed by the tendency of neuroscientists to make big, bold claims about disease mechanisms based on studies with tiny sample sizes. Both sides make reasonable criticisms. As I said in the quote from my previous article above, it seems likely that disease heterogeneity - that is, multiple diseases states with the same broad end point being simplistically lumped together - plays a major role in the failure of genome-wide association studies of psychiatric conditions; at the same time, the scientific value of much of the "sexy" neurobiology currently being published (e.g. functional MRI finds that conservatives have lower activity in "compassion" centres of the brain, or whatever) is sometimes highly questionable. Both sides of this scuffle have something to learn from their opponents. The editorial argues, sensibly, that geneticists and neuroscientists just need to start getting along. The ideal situation is one in which rigorous clinical assessments are used to generate patient cohorts that are as homogeneous as possible that can then be subjected to large-scale genetic analysis. One especially promising avenue is the use of "endophenotypes" - that is, simple and easily quantifiable traits that are sometimes but not always associated with a particular disease. Cleanly defined endophenotypes, such as very specific dysfunctions of brain activity, may prove much more amenable to genetic dissection than the larger, more complex diseases they are associated with. Comprehensively tackling the genetic of psychiatric conditions will require a forceful and combined approach drawing on the clinical expertise of neuropsychiatrists and the experience of geneticists in unravelling the genetic mechanisms of complex traits. To some extent this is happening already (no large genetics consortium would be naive enough to embark on a multi-million dollar project without consulting clinical experts) - but obviously there is considerable room for improvement. Moving beyond common SNPs  Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation. Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation.The approaches required to capture these variants are already pretty well-known, although they remain expensive and technically challenging. In an ideal world, genome-wide association studies would be truly genome-wide - in other words, they would utilise the entire DNA sequence of all of the patients and controls in the sample to find every possible genetic variant that might contribute to disease. Unfortunately, such an approach is currently out of reach, for several reasons:
Both approaches have their limitations. The success of the candidate gene approach will be constrained by researchers' ability to identify the genes most likely to be involved in a particular disease - but in fact our currently severaly limited understanding of disease genetics is precisely why we need to study this issue in the first place! (In the Nature news piece, Harvard's Steven Hyman memorably describes this approach as "like packing your own lunch box and then looking in the box to see what's in it.") And while chip-based detection of structural variation is rapidly increasing in resolution, it's extremely difficult to determine which of the variants identified in a study are disease-causing and which are harmless polymorphisms - this is currently done probabilistically, by showing that there is an enrichment of new variants in disease cases compared to controls, but this approach cannot tell you which of the identified variants are actually causative. From psychiatric genetics to genetic psychiatry? There are several important reasons researchers are interested in the genetics of mental illness: identifying causal genes helps to dissect out the molecular pathways involved in disease, and may help to pull out otherwise invisible sub-types of a disease; studying "extreme" mental phenotypes may illuminate the genetic basis of variation in cognition and personality traits in "normal" people; and, perhaps most importantly, by identifying the genes underlying psychiatric diseases we may be able to target at-risk individuals for monitoring and intervention, potentially heading off severe disease before it takes hold. In the headlong pursuit of these goals the field of psychiatric genetics has developed an unfortunate reputation built on bold claims made with limited evidence, and literally hundreds of reported associations that have completely failed to stand up to replication. Just a couple of years ago the shiny new tools of large-scale genomics promised an end to this ignoble period in the history of the field; unfortunately, the introduction of larger samples, higher genomic coverage and increased statistical rigour has not brought the desired clarity to the field, but rather seems to have increased the levels of confusion and uncertainty. If anything, that crucial third goal - using genetic to predict the risk of mental illness - now appears further away than it did just a couple of years ago. Back in early 2007 we didn't have many convincing genetic predictors of mental illness, but at least it was possible to imagine that emerging genomic technologies might identify a small core set of large-effect variants that would help clinicians to predict disease risk. Right now we still don't have many useful genetic predictors, and that illusion of hope is gone. In summary: while there's no doubt that these conditions do have a strong genetic basis, it's now abundantly clear that this basis is frighteningly complex, with common variants of moderate-to-large effect - the types of variants that would be most useful for risk prediction - being essentially absent. It's going to take many years, massive cohorts, the clever application of new genomic technologies, and a willingness from both neuroscientists and geneticists to listen to one another to move this field forward. (Brain scan image from Science Photo Library.) * Thanks to reader Chris for providing me with the citation, which I had carelessly misplaced! |
| Misha Angrist reviews personal genomics [Genetic Future] Posted: 19 Aug 2008 09:38 AM CDT Blogger, Personal Genome Project participant and Assistant Professor Misha Angrist has a concise and extraordinarily readable article on the current state of personal genomics at Technology Review. Here's the penultimate paragraph: This is where we are in the era of personal genomics: some modest amusement, a few interesting tidbits, a bit of useful information, but mostly the promise of much better things to come. The more people are allowed--encouraged, even--to experiment, the sooner that promise can be realized.I find myself in complete agreement. Anyone interested in the field should go read the rest. |
| Posted: 19 Aug 2008 09:05 AM CDT Nowadays, as many folks peer into the vast tangled thicket of their own genetic code, they, as I, assuredly wonder what it all means and how best to ascertain their health risks. One core theme that emerges from repeated forays into one’s own data is that many of us carry a scads of genetic risk for illness, but somehow, find ourselves living rather normal, healthy lives. How can this be ? A recent example of this entails a C/T snp (rs35753505) located in the 5′ flanking region of the neuregulin 1 gene which has been repeatedly associated with schizophrenia. Axel Krug and colleagues recently reported in their paper, “Genetic variation in the schizophrenia-risk gene neuregulin1 correlates with differences in frontal brain activation in a working memory task in healthy individuals” that T/C variation at this snp is associated with activation of the frontal cortex in healthy individuals. Participants were asked to keep track of a series of events and respond to a particular event that happened “2 events ago” . These so-called n-back tasks are not easy for healthy folks, and demand a lot of mental focus - a neural process that depends heavily on circuits in the frontal cortex. Generally speaking, as the task becomes harder, more activity in the frontal cortex is needed to keep up. In this case, individuals with the TT genotype seemed to perform the task while using somewhat less activity in the frontal cortex, rather than the risk-bearing CC carriers. As someone who has tried and failed to succeed at these tasks many times before, I was sure I would be a CC, but the 23andMe data show me to be a non-risk carrying TT. Hmmm … maybe my frontal cortex is just underactive. |
| 4 Million for Education [The Gene Sherpa: Personalized Medicine and You] Posted: 19 Aug 2008 08:50 AM CDT |
| Linux is so not ready for the desktop [Mailund on the Internet] Posted: 19 Aug 2008 12:33 AM CDT …or maybe I am just not ready to use it there… Actually, I have been using Linux for more than 10 years and, until I bought a Mac this year, exclusively since 1995. Generally, I am very comfortable with Linux, but I made a major mistake when I bought my latest office machine about a year ago. I bought a Dell ’cause I figured that since Dell sells machines with Ubuntu pre-installed (in the US and UK but sadly not in Denmark) the hardware would be supported by Linux. Not so! This machine has an ATI Radeon HD graphic card, and Linux support for that is practically non-existing. ATI has a driver for it that you can install, and with that I managed to get an acceptable — but by no means very good — performance. 2D graphics at least. 3D graphics rarely worked. I begged Kristian Høgsberg for help. He is a friend of mine who works at RedHat, making eye candy for Linux. I figured he would have some suggestions. He suggested I switched from Ubuntu to Fedora, ’cause he wouldn’t know how to help me with a competing product. I figured he was joking, but still, I switched. In Fedora, I actually managed to get the ATI driver working. Not perfectly, but a lot better than I had managed before. Now, a few days ago, it stopped working. I’m not sure what happened. I didn’t fiddle around with the setup or any thing, but maybe updated package or something broke the setup. Break it did, to the point where enabled Compiz meant the screen would just gray out and where disabled Compiz was painfully slow. Like you wouldn’t imagine. Moving a window across the screen would take several seconds. In frustration and desperation I re-installed Fedora yesterday. I was hoping I would get it back to the state it was a few days ago. That didn’t work, but I did manage to delete a week’s work that I hadn’t backed up. Way to go, Dr. Mailund, way to go! I caught Høgsberg on Jabber and asked for help again. He suggested that I buy a nVidea graphics card. That is probably the right choice. Anyway, I don’t have time to try to fix my Linux. I need to analyse some data before a meeting in Iceland next week and I’m leaving Friday. I need Linux for it, though, ’cause I haven’t managed to compile the software I need on my Mac. I guess I’ll just ssh to the Linux box and work that way. |
| A service that I might use [business|bytes|genes|molecules] Posted: 19 Aug 2008 12:04 AM CDT CNET reports that Khosla Ventures has led an investment round of $3 million in ZocDoc, a health 2.0 company a little off the beaten path. ZocDoc, which currently operates only in Brooklyn and Manhattan, allows you to find an appointment with a doctor or dentist. Now that’s a service I could really use. It matches doctors to insurance and a rating system. I don’t know how well it works since I really can’t make good use of it living in Seattle, but the company, which is about a year old, is providing the kind of simple service that could be really useful if done right. Related articles by Zemanta |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |
 This is an exciting time in the study of hereditary factors involved in breast cancer susceptibility. Breast cancer for a long time was classified according histology. Now genetics play a significant role and better knowledge ensures better management and treatment.
This is an exciting time in the study of hereditary factors involved in breast cancer susceptibility. Breast cancer for a long time was classified according histology. Now genetics play a significant role and better knowledge ensures better management and treatment.


![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_b.png?x-id=d030c1ac-1feb-45d3-b407-1e99b8a1a63c)
No comments:
Post a Comment