The DNA Network |
| The challenges of psychiatric genetics [Genetic Future] Posted: 06 Aug 2008 06:57 PM CDT  Back in April I posted on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year. Back in April I posted on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year.The major message from that article is that although bipolar disorder is massively influenced by genetic factors (around 85% of the variation in risk is thought to be due to genetics) we still don't really have the faintest idea exactly which genes are involved. This is despite three reasonably large genome-wide association studies involving over 4,000 bipolar patients in total, which generated weak and contradictory results and failed to provide a single compelling candidate for genetic variation underlying this disease. This disappointing result has also held largely true for other psychiatric conditions with strong genetic components, such as schizophrenia, major depression and autism. Genetic studies of these conditions have had some success identifying rare mutations that underlie severe cases, but the vast majority of the genetic variants contributing to risk remain undiscovered. There are several reasons why genome-wide association studies can fail to yield significant harvests of disease-associated genes. I summed these up with respect to bipolar disease as follows: The researchers are surely hoping that small effect sizes are the major problem, since this is the easiest problem to remedy (simply increase sample sizes). Disease heterogeneity - in other words, multiple diseases with distinct causes that all converge on a bipolar end-point - also seems like a particularly plausible explanation given the complexities of mental illness. It's also likely that various types of genetic variants that are largely invisible to existing SNP chips, like rare variants and copy-number variation, are important.The same story probably holds largely true for other psychiatric conditions. In this week's issue of Nature, a news article and an editorial both tackle the challenges of psychiatric genetics, and lay out the ambitious strategies currently being pursued by researchers around the world to overcome them. Small effect sizes The first hurdle that I describe above is the fact that most of the variants underlying these conditions probably have very small effect sizes (only increasing risk by less than 20%). Such variants will only be identified by cranking up sample sizes immensely, an approach that has yielded some limited success for other genetically complex traits such as height and obesity. The Nature news feature has a table listing some of the major collaborative efforts currently collecting genetic information from the very large cohorts required to dissect out the basis of these conditions: 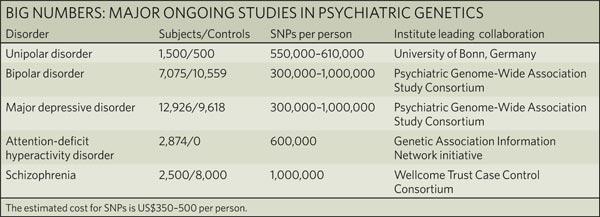 In most cases, these samples are being built up by pooling results from multiple different studies, often gathered by groups from around the world. As sample sizes increase the power of studies to detect small-effect variants grows. The effect of sample size on the power of genome-wide association studies is illustrated in the graph below (which I've managed to save without recording a source - if anyone recognises it, please let me know): 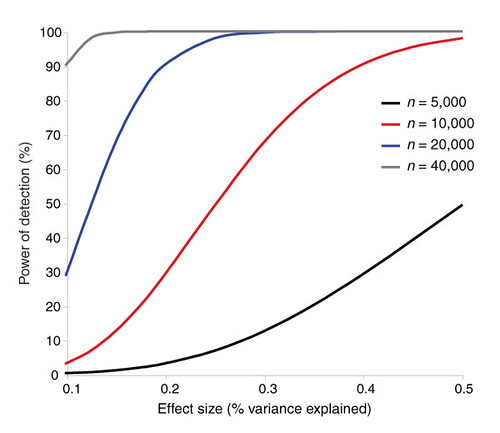 Take a single genetic variant that explains just 0.5% of the variance in the risk of a psychiatric disorder. With a sample size of 5,000 individuals with that disorder you still have a mere 50% chance of detecting that variant. Double your sample size, and that probability jumps to a near-certainty of detection - and your power of detecting even smaller-effect variants (explaining, say, 0.2% of the risk) starts to climb to respectable levels. By staring at those curves for a while, and bearing in mind that many of the variants found by recent genome-wide association studies explain well under 0.2% of the risk variance, you will quickly start to appreciate why researchers are pushing for ever-larger disease populations to work with. With truly enormous samples on the order of 50 to 100 thousand patients - not out of the question for international consortiums studying reasonably common diseases such as bipolar - the power to detect even very weak risk variants becomes reasonable. If there are common genetic variants contributing to the risk of these diseases, such large collaborative studies will eventually find them; so long, of course, as they can tackle the next (and potentially far more serious) problem of disease heterogeneity. Complex, heterogeneous diseases  The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible. The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible.This complexity and heterogeneity is the basis of considerable tension between geneticists and neuroscientists, which is explored in the Nature editorial. Basically, to build up those massive sample sizes shown above geneticists are forced to lump together patients with a variety of clinical symptoms, thus essentially ignoring the complexity inherent in these conditions - a failure that neuroscientists find inexcusable. In turn, geneticists (like myself) get seriously annoyed by the tendency of neuroscientists to make big, bold claims about disease mechanisms based on studies with tiny sample sizes. Both sides make reasonable criticisms. As I said in the quote from my previous article above, it seems likely that disease heterogeneity - that is, multiple diseases states with the same broad end point being simplistically lumped together - plays a major role in the failure of genome-wide association studies of psychiatric conditions; at the same time, the scientific value of much of the "sexy" neurobiology currently being published (e.g. functional MRI finds that conservatives have lower activity in "compassion" centres of the brain, or whatever) is sometimes highly questionable. Both sides of this scuffle have something to learn from their opponents. The editorial argues, sensibly, that geneticists and neuroscientists just need to start getting along. The ideal situation is one in which rigorous clinical assessments are used to generate patient cohorts that are as homogeneous as possible that can then be subjected to large-scale genetic analysis. One especially promising avenue is the use of "endophenotypes" - that is, simple and easily quantifiable traits that are sometimes but not always associated with a particular disease. Cleanly defined endophenotypes, such as very specific dysfunctions of brain activity, may prove much more amenable to genetic dissection than the larger, more complex diseases they are associated with. Comprehensively tackling the genetic of psychiatric conditions will require a forceful and combined approach drawing on the clinical expertise of neuropsychiatrists and the experience of geneticists in unravelling the genetic mechanisms of complex traits. To some extent this is happening already (no large genetics consortium would be naive enough to embark on a multi-million dollar project without consulting clinical experts) - but obviously there is considerable room for improvement. Moving beyond common SNPs  Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation. Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation.The approaches required to capture these variants are already pretty well-known, although they remain expensive and technically challenging. In an ideal world, genome-wide association studies would be truly genome-wide - in other words, they would utilise the entire DNA sequence of all of the patients and controls in the sample to find every possible genetic variant that might contribute to disease. Unfortunately, such an approach is currently out of reach, for several reasons:
Both approaches have their limitations. The success of the candidate gene approach will be constrained by researchers' ability to identify the genes most likely to be involved in a particular disease - but in fact our currently severaly limited understanding of disease genetics is precisely why we need to study this issue in the first place! (In the Nature news piece, Harvard's Steven Hyman memorably describes this approach as "like packing your own lunch box and then looking in the box to see what's in it.") And while chip-based detection of structural variation is rapidly increasing in resolution, it's extremely difficult to determine which of the variants identified in a study are disease-causing and which are harmless polymorphisms - this is currently done probabilistically, by showing that there is an enrichment of new variants in disease cases compared to controls, but this approach cannot tell you which of the identified variants are actually causative. From psychiatric genetics to genetic psychiatry? There are several important reasons researchers are interested in the genetics of mental illness: identifying causal genes helps to dissect out the molecular pathways involved in disease, and may help to pull out otherwise invisible sub-types of a disease; studying "extreme" mental phenotypes may illuminate the genetic basis of variation in cognition and personality traits in "normal" people; and, perhaps most importantly, by identifying the genes underlying psychiatric diseases we may be able to target at-risk individuals for monitoring and intervention, potentially heading off severe disease before it takes hold. In the headlong pursuit of these goals the field of psychiatric genetics has developed an unfortunate reputation built on bold claims made with limited evidence, and literally hundreds of reported associations that have completely failed to stand up to replication. Just a couple of years ago the shiny new tools of large-scale genomics promised an end to this ignoble period in the history of the field; unfortunately, the introduction of larger samples, higher genomic coverage and increased statistical rigour has not brought the desired clarity to the field, but rather seems to have increased the levels of confusion and uncertainty. If anything, that crucial third goal - using genetic to predict the risk of mental illness - now appears further away than it did just a couple of years ago. Back in early 2007 we didn't have many convincing genetic predictors of mental illness, but at least it was possible to imagine that emerging genomic technologies might identify a small core set of large-effect variants that would help clinicians to predict disease risk. Right now we still don't have many useful genetic predictors, and that illusion of hope is gone. In summary: while there's no doubt that these conditions do have a strong genetic basis, it's now abundantly clear that this basis is frighteningly complex, with common variants of moderate-to-large effect - the types of variants that would be most useful for risk prediction - being essentially absent. It's going to take many years, massive cohorts, the clever application of new genomic technologies, and a willingness from both neuroscientists and geneticists to listen to one another to move this field forward. (Brain scan image from Science Photo Library.) |
| Writing for informavores [Genetic Future] Posted: 06 Aug 2008 06:55 PM CDT Slate has a great article about the way people read online. Apparently the typical internet reader doesn't tolerate big slabs of uninterrupted text, so I should really be writing fewer posts like this one, and more posts like the one you're reading now. |
| Support animal research, save lives [Discovering Biology in a Digital World] Posted: 06 Aug 2008 06:21 PM CDT When female bloggers get death threats for comparing a Batman movie to a poor business plan, and friends can have their lab fire bombed for doing plant genetics, it's sometimes a little scary to step into the fray and take a stand on controversial issues. But that's the point. We have to speak out. Scary or not, unless we speak out against the animal rights terrorists who firebomb people's homes and harass researchers, we will lose any chance to save our loved ones from diseases like cancer, HIV, Alzheimer's, or many others. Let's think about just one of these. Read the rest of this post... | Read the comments on this post... |
| Should Well-Educated People Know Math and Science? [adaptivecomplexity's column] Posted: 06 Aug 2008 05:18 PM CDT A physics professor, writing in Inside Higher Ed, asks why intellectuals think it's ok to be ignorant of math and science, but not of art, music and literature. When among intellectual company, humanities professors can confess, without a trace of shame, their complete ignorance of science, one of humanity's most important intellectual achievements. But in our culture, a science professor had better not admit to a similar level of ignorance about art or music. This physics professor quotes another blogger to illustrate the phenomenon:
To which he replies: |
| Tomorrow's Table in the classroom [Tomorrow's Table] Posted: 06 Aug 2008 04:58 PM CDT "I really enjoyed the book. It did a great job of keeping everything in perspective. Use again !" "Use again! A great resource and easy to understand" "The textbook was great. It had a story line to it. It was easy to remember." These are some of the comments from Oregon State University students who read the book, "Tomorrow's Table: Organic Farming, Genetics and the Future of Food". Steven Strauss, Distinguished Professor of Forest Biotechnology at Oregon State University, who directs the OSU Program for Outreach in Resource Biotechnology, chose the book for his course, which give students and the public scientifically reliable information about the use of genes and chemicals in agriculture and natural resources. Thanks Steve, for being the first to use it in the classroom! |
| Just give us the eggs...women's health be damned [Mary Meets Dolly] Posted: 06 Aug 2008 12:52 PM CDT I read this article and it absolutely enraged me. Dr. Sam Wood of Stemagen Corp. wants to pay women to harvest their eggs. He cries that he can't get them any other way. Forget about the health risks to young women, Dr. Wood wants the eggs to continue to pursue therapeutic cloning. It is all about the money. From CNNMoney.com:
Well, sign me up! If Dr. Wood is so convinced that my eggs can change the world. But wait, let us ask ourselves why Dr. Wood is so adamant that therapeutic cloning is the answer even though there is no data to support his claim. The California agency that doles out the 3 billion dollars in taxpayer money for stem cell research has denied all requests for funding of therapeutic cloning:
So Dr. Wood can't get taxpayer money without an abundant supply of eggs, but he can't get that without paying young women to put their health, their fertility and possibly their lives at risk. Therapeutic cloning to obtain human stem cells is barely a reality and Dr. Wood wants thousands of women to donate to the cause. Dare I say it: The unmitigated arrogance of some people! Talk about treating women as objects. I think Dr. Wood may out-objectify the porn industry. I will leave you with Dr. Wood's most illuminating last words: "Give us the eggs. If we don't succeed, then be critical," said Wood. "You have to give people the tools that are required to determine whether the methodology will work." |
| Please Don't deactivate me! [The Gene Sherpa: Personalized Medicine and You] Posted: 06 Aug 2008 12:34 PM CDT |
| Thanks Carl, you're ruined it for me forever [Discovering Biology in a Digital World] Posted: 06 Aug 2008 11:00 AM CDT Microbiologist develop some strange habits when it comes to food. Some take a fatalistic approach. They reason that microbes are everywhere, we're going to die anyway, we might as well eat dirt and make antibodies. You know these people. They quote things like the "10 second rule" when food drops on the floor, tell you we're all getting asthma because we're too obsessed with cleanliness, and let their dogs wash their dishes. Eeew. With a few possible exceptions, I'm in the other camp. Read the rest of this post... | Read the comments on this post... |
| Labmeeting: It really makes science easier [ScienceRoll] Posted: 06 Aug 2008 09:58 AM CDT I’ve written many posts about community sites created for scientists and medical professionals, but I do not really prefer any of them as they are quite similarly constructed. Labmeeting will be the first service I plan to use actively. Why? Upload research articles
Browse different streams of new papers
Manage a lab webspace
 |
| Simple Genetics: Wiki of genetic test reviews [ScienceRoll] Posted: 06 Aug 2008 09:21 AM CDT This site has been on my list for a while.
Though, the site seems to be inactive and we all know how much chance a new wiki has nowadays to become a well-edited, regularly updated database. I tell you, not much… (Hat Tip: Cancer Genetics)  |
| Do you think medical education can be reformed? How? [ScienceRoll] Posted: 06 Aug 2008 08:56 AM CDT This is a question I asked on the Medical Education Evolution community page. Here are some interesting answers from famous educators and medical professionals:
Others think differently:
Further reading:  |
| Posted: 06 Aug 2008 08:30 AM CDT This the third part of case study where we see what happens when high school students clone and sequence genomic plant DNA. In this last part, we use the results from an automated comparison program to determine if the students cloned any genes at all and, if so, which genes were cloned. (You can also read part I and part II.) Did they clone or not clone? That is the question. Read the rest of this post... | Read the comments on this post... |
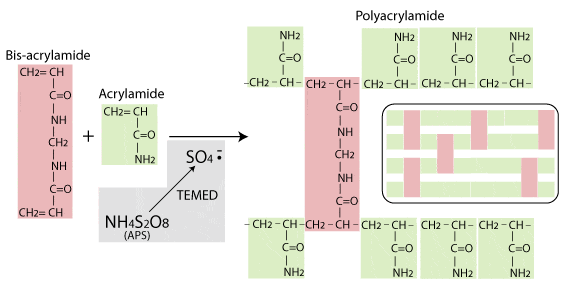
| Agarose Gels Do Not Polymerise! [Bitesize Bio] Posted: 06 Aug 2008 06:24 AM CDT I was browsing a certain website the other day when I came across a protocol that advised: “When agarose starts to cool, it undergoes what is known as polymerization” Polymerising agarose? Someone is getting their gel matrices mixed up, and it’s not the first time I have heard this said, so it looks like some myth-busting is required. So hunker down for a quick run down on the difference between polymerising and non-polymerising gel matrices. Of the common gel matrices used in molecular biology, polyacrylamide, agar and agarose, polyacrylamide is the one that polymerises. I’ll deal with agarose and agar later. Polyacrylamide gel polymerisation Polyacrylamide, used mainly for SDS-PAGE, is a matrix formed from monomers of acrylamide and bis-acrylamide. It’s strengths are that is it chemically inert - so won’t interact with proteins as they pass through - and that it can easily and reproducibly be made with different pore sizes to produce gels with different separation properties. The polymerisation reaction, shown in the diagram below, is a vinyl addition catalysed by free radicals. The reaction is initiated by TEMED, which induces free radical formation from ammonium persulphate (APS). The free radicals transfer electrons to the acrylamide/bisacrylamide monomers, radicalizing them and causing them to react with each other to form the polyacrylamide chain. In the absence of bis-acrylamide, the acrylamide would polymerise into long strands, not a porous gel. But as the diagram shows, bis-acrylamide cross-links the acrylamide chains and this is what gives rise to the formation of the porous gel matrix. The amount of crosslinking, and therefore the pore size and consequent separation properties of the gel can be controlled by varying the ratio of acrylamide to bis-acrylamide.
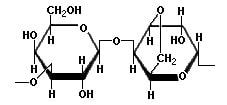
For more information on polyacrylamide gel polymerisation see Biorad Bulletin 1156 Agarose gel formation So what about agarose? Well, agarose - the main component of the gelatinous agar that can be isolated from certain species of seaweed - is itself a polymer. But, polymerisation is not the mechanism for agarose gel formation. Chemically, agarose is a polysaccharide, whose monomeric unit is a disaccharide of D-galactose and 3,6-anhydro-L-galactopyranose which is shown in the diagram below.
In aqueous solutions below 35°C these polymer strands are held together in a porous gel structure by non-covalent interactions like hydrogen bonds and electrostatic interactions. Heating the solution breaks these non-covalent interactions and separates the strands. Then as the solution cools, these non-covalent interactions are re-established and the gel forms. So agarose (and agar) gels form by gellation through hydrogen bonding and electrostatic interactions, not through polymerisation |
| The Radar is looking at some bio geek topics [business|bytes|genes|molecules] Posted: 05 Aug 2008 11:28 PM CDT The O’Reilly Radar is tracking some themes of interest to them, and all relevant to our bio geek community Synthetic Biology I’ve had some disagreements with the tech geek community’s understanding of the life sciences, so will be interesting to see what tack the Radar takes. I also suspect they will all be covered at this years Scifoo Related articles by Zemanta |
| Is your web service open source? [business|bytes|genes|molecules] Posted: 05 Aug 2008 10:06 PM CDT
Yesterday, on Friendfeed, Aarthy asked the following question
I’d like to point to the bold part. It’s along the lines of a topic that Tim O’Reilly has talked about over the years and revisited after OSCON. I have heard a few discussions recently where people are trying to choose between making software source available to academics (not necessarily the same as open source), making software open source and/or providing a web service. The thing that jumps out at me is that people seem to be confused. They don’t always know why they want to make software available a certain way, or what the pros and cons of the various options are. So the question I have is this. Let us say there is a service that provides certain functionality, let’s say it allows you to do phylogenetic analysis of DNA sequences. Would most users be satisfied with a high quality web service with programmatic access for those who want to use the service in conjunction with other tools? Would users be satisfied with an associated publication or application note describing the underlying science? Or would you want a source code download under all circumstances? My personal preference, in most cases, is to provide a web service with an open, well documented, API, make sure that the algorithm is described somewhere, and open source the algorithm, or at least the original implementation (in this case the GPL is a pretty good idea) . In today’s world, that means, put the source up on Sourceforge, Github, or Google Code. Of course, there is a challenge here. How do you monetize if you want to? I am somewhat conflicted here. In the commercial open source world, the success stories (Canonical, RedHat, MySQL, etc) have been successful partly due to the volume of users and partly due to the ability of those efforts to become a core part of the business processes of a number of companies. There are very few applications in the life sciences that fall into that category, and none of them are infrastructure applications. I’ve been around long enough to recognize that many people, especially those in the biopharma industry like the idea of paying for customer support, or having a rich molecular graphics environment around well known algorithms, so there is value to commercialization, including open source software. On the other hand, companies like Google have been built on top of open source software but, e.g. in Google’s case, the engine that makes them tick is often proprietary (although in some cases certain aspects are published). So where does life science software fall? First of all, we should end the sham that is academic-only software. It belongs to a different era. In many cases, a small VC-funded biotech has less money to spend on software than most academic labs. Make the software open source, in the proper way. If you want to make money from your work, form a compelling partnership, or in a world with virtual resources, encourage people in the lab to develop a commercial service. Give people something they care about and will find real value from. If you are a company, you need to publish your algorithm and perhaps open source the original implementation. I do believe in trade secrets in the appropriate situation, but the number of algorithms out there which might merit that kind of protection are few and far between. In the end, much like data, it’s the implementation, support of the implementation and value add that makes a particular software package worth paying a subscription for. Which brings me back to the quote that started all of this. Would love to hear your thoughts, cause it’s something we need to think about in a rapidly changing computing environment. Prologue: I mentioned the GPL earlier in this text. Over the past few years, I have somewhat soured on copy-left licensing. IMO, there is place for the GPL, for example a complete software suite, or a core algorithm (the example above). However, for a big chunk of cases, e.g. a library of some kind, I much prefer more permissive licenses, e.g. BSD, MIT, Apache. If someone can innovate based on what you might have developed it’s a good thing. If that person is you, even better. Update: Updated the part about “there is no such thing as an open source web service. As Andrew Clegg pointed out on Friendfeed, you can make the entire source of a web service available. That leads to the follow up question in my head. In a world where your computational resources are more and more virtual, what’s the point of an open source web service? That will be the discussion point of a follow up post Related articles by Zemanta
|
| Posted: 05 Aug 2008 08:37 PM CDT Physicist Nobel Laureate Philip Anderson on computers and physics:
(That last part should be qualified: simulations can prove things, but those things may have no resemblance to what goes on in the physical world.) In this short piece, Anderson argues that in physics, the tremendous power of computers is more helpful in organizing experimental data than in guiding theorists. Simulations of complex systems that try to account for the interactions of every single atom involved are out of reach of our current computer power, and the shortcuts that simulators make to get around this limitation end up severely biasing the results of the simulation. Thus theorists, Anderson argues, should not be using simulations as a substitute for "logic and pure science". |
| Interesting Genomic Tools [ScienceRoll] Posted: 05 Aug 2008 10:21 AM CDT I’ve recently come across some great genomic tools that may be useful for your research.
If you know any more, don’t hesitate to share those with us.  |
| Fighting cancer with video games: Conclusion [ScienceRoll] Posted: 05 Aug 2008 09:35 AM CDT Do you remember my post about Hopelab and their video game constructed to help children fighting cancer? Now an article was published in Pediatrics about the outcomes and advantages of using such video games in treating patients. An excerpt from the study:
Further reading:  |
| Medicine 2.0 Microcarnival: Launch! [ScienceRoll] Posted: 05 Aug 2008 08:48 AM CDT I told you about my problems with blog carnivals before. I will update and manage all of my carnivals, but I will also launch a microcarnival on Friendfeed to see whether it works or not. Join that Friendfeed room if you are interested to read web 2.0/medicine related news and blogposts.  |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |









No comments:
Post a Comment