The DNA Network |
| On using "information processing" as a metaphor for biological processes [The Daily Transcript] Posted: 12 Aug 2008 08:52 PM CDT Sorry, I've been busy these past few days. I want to respond to a comment posted by Dan and take this opportunity to broaden the discussion about how we use language to construct models. Dan's concerns about information and life have been echoed by many out there, for example by John Wilkins. Can we use the term "information" when discussing life? Is there such a thing as "information"? Are these buzzwords without any deeper meaning? What is lost in such an analysis is that all of our theories are infused with metaphors. These words and concepts help us to better understand the ideas and insights that may come out of any particular model. The metaphor of information and the use of similar terms in molecular biology (signal transduction, transmit, secondary messenger, genetic program, mRNA, transcription, translation, copying, amplification) help us to comprehend underlying biological processes. A certain sequence of base pairs is passed on from generation to generation, information. A cell secrets a growth factor and causes a nearby cell to differentiate, information. Does information exist per say? Well I would like to argue that nothing exist in the exact way that any term implies, words after all are simply tools that help us to understand the world in which we live in. Some words are better than others, some less so. Some words give more insight because their meanings give insight when invoked in a particular context. You may choose to eliminate all the words for information in your explanation of biological processes and use instead another metaphor, that of machines for example, and you may gain some different insight, but let's face it, you will not be able to easily understand all the issues and questions that are implicit in the study of classical genetics. What is the unit of heredity? How is genetic information transmitted? Does genetic information have a physical counterpart? How is the sequence of bases along a strand of DNA converted into a sequence of amino acids? To say that information is a buzz word, or that it doesn't exist misses the point. It is a useful analogy. It's a metaphor that gives insight, just like the idea of molecular motors can promote the development of other forms of deeper understanding. Read the rest of this post... | Read the comments on this post... |
| If you really want to help animals... [Discovering Biology in a Digital World] Posted: 12 Aug 2008 08:36 PM CDT write your Senators and Representatives about saving the Endangered Species Act. But, first read what Mike Dunford has to say. Mike describes the changes that the Bush administration has proposed in great detail and consequences for wild animals. Greg Laden has posted on this, too. Read the comments on this post... |
| Hail To The Red, White And Green [] Posted: 12 Aug 2008 07:08 PM CDT
Okay, maybe this is obvious to everybody else by now, but we’ve just run across the red, white and green differentiations for biotech. Red biotechnology refers to transgenic microorganisms for use in the production of pharmaceutical and medical substances; White biotechnology leverages transgenic organisms — usually moulds, enzymes, yeast and bacteria – to produce industrial chemicals; Green biotechnology involves the creation of transgenic crops to increase crop yields (by resistance to pests and disease, immunity to herbicides, temperature extremes, etc.). |
| Who's your doggie's daddy? Ancestry testing for dogs [Discovering Biology in a Digital World] Posted: 12 Aug 2008 06:54 PM CDT Ancestry tests aren't just for humans anymore. We went to Petco this weekend to buy dog food and found brochures for doggy DNA testing. Now, those of you with dogs of uncertain parentage need puzzle no longer. According to Petco, their SNP test (what is a SNP?) can identify over 100 different breeds and they'll tell you which breeds are represented in your dog and whether your dog's breeding is mixed (or pure). Read the rest of this post... | Read the comments on this post... |
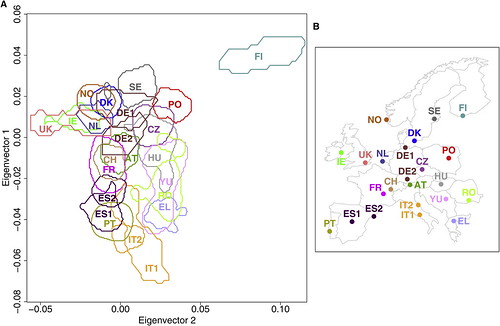
| How well does your genome predict your postcode? [Genetic Future] Posted: 12 Aug 2008 06:21 PM CDT Well, it's far from GPS precision, but the concordance between this genetic map of Europe (below left) and the physical sampling locations of populations throughout Europe (below right) is pretty good for a first draft:
Dienekes has an excellent discussion of the technical details, while Razib has labelled a plot showing all of the individuals in the study to make it easier to assess the degree of scatter and overlap. The take-home message: rather than being one homogeneous mass, Europeans in fact show considerable population substructure, such that genetic information can be used to roughly predict geographical ancestry. An analysis of just a few hundred thousand genetic markers (i.e. less than is currently offered by personal genomics companies 23andMe or deCODEme) would be more than adequate in most cases to distinguish a Pole from a Parisian, or a Swede from a Spaniard. (To be more precise, it would be sufficient to discriminate between individuals for whom most ancestors were natives of these regions; recent migrants will obviously be misclassified.) What drove these genetic differences? Mostly it will have been chance - random increases or decreases in the frequency of markers throughout the genome accumulated over a few millennia of genetic isolation. But at least some of these differences have been driven by natural selection: for instance, the lactase gene LCT, which has been subject to strong selection to allow lactose digestion in adults in populations reliant on dairy agriculture, represents 9 out of the top 20 most differentiated markers; a marker in the gene HERC2, which is associated with eye colour variation and has been under selection in Europeans and Asians, comes in at number 19. This indicates that at least some of the genetic - and thus physical and possibly behavioural - differences between the various European populations stem from evolutionary adaptation to their local environments. I'll leave the technical commentary to Dienekes, but I do want to make one important point: the accuracy of the map will have been limited by the fact that the markers used in this study represent sites of common variation; data from large-scale genome sequencing will generate far, far better maps. The major reason for this is that sequencing will provide information on rare, highly spatially-restricted variants - many of which will be limited to single families and thus be extremely informative about geographical ancestry. Basically, if you had complete genome sequences from enough Europeans you could reconstruct the genetic map of Europe with exquisite precision. In addition to empowering genetic genealogists, researchers could use deviations between the genetic and physical maps to make powerful inferences about historical migration events and recent episodes of natural selection. With any luck, this is the sort of data that will simply fall out from large-scale population genomic studies being conducted over the next decade or so. Update: Kambiz at Anthropology.net puts these results in a broader scientific context. Lao et al. (2008). Correlation between Genetic and Geographic Structure in Europe. Current Biology DOI: 10.1016/j.cub.2008.07.049 |
| Large reservoir of mitochondrial DNA mutations identified in humans [Think Gene] Posted: 12 Aug 2008 06:06 PM CDT Josh: We must remember that many genetic diseases are not caused by mutations in nuclear DNA, but are the result of mutations in mitochondrial DNA. The press release makes mention that the number of cells with mitochondrial mutations often determines the severity of disease; I assume this refers to the ratio of normal:mutant mitochondria within a cell. Remember, the egg had more than a single mitochondrion, and they may or may not all be genetically identical. When more mitochondria are produced in a cell, they may not all replicate at an even rate. Researchers at the University of Newcastle, England, and the Virginia Bioinformatics Institute at Virginia Tech in the United States have revealed a large reservoir of mitochondrial DNA mutations present in the general population. Clinical analysis of blood samples from almost 3,000 infants born in north Cumbria, England, showed that at least 1 in 200 individuals in the general public harbor mitochondrial DNA mutations that may lead to disease. The findings, which highlight the need to develop new approaches to prevent the transmission of mitochondrial diseases, were published in The American Journal of Human Genetics. Mitochondria, the “engines” present in each cell that produce adenosine triphosphate, are passed from mother to offspring. Mutations in mitochondrial DNA inherited from the mother may cause mitochondrial diseases that include muscle weakness, diabetes, stroke, heart failure, or epilepsy. In almost all mitochondrial diseases caused by mutant mitochondrial DNA, the patient’s cells will contain a mixture of mutant and normal mitochondrial DNA. The proportion of mutant mitochondrial DNA in most cases determines the severity of disease. Previous estimates from epidemiological studies suggested that mitochondrial diseases affect as many as one person in 5,000. However, the incidence of new mitochondrial mutations and the prevalence of those carrying these mutations were never fully established due to limitations in the methods used. Most of the earlier estimates of the frequency of mitochondrial DNA mutations in the general population, for example, have depended on identification of clinically affected patients and subsequent retracing of inheritance on the maternal side of the family. This approach fails to detect the gradual accumulation of mutations in some members of the population, including those individuals who harbor mitochondrial DNA mutations but who otherwise do not show the symptoms of disease. Dr. David Samuels, Assistant Professor at the Virginia Bioinformatics Institute and an author on this study, commented: “We know from many clinical studies of patients and their families that our cells can tolerate a rather large amount of mutant mitochondrial DNA with no significant loss of function. From that observation we have suspected that there may be a large number of people in the general population who carry pathogenic mitochondrial DNA mutations, but who are not obviously ill with a mitochondrial disease. This study gives us, for the first time, a measurement of the number of these carriers of pathogenic mitochondrial DNA mutations in the general population. One in every 200 individuals is a lot of people – around 1.5 million people in the United States alone. ” The scientists looked at 10 mitochondrial DNA mutations (arising from single nucleotide replacements) often found in patients with mitochondrial disease. By taking advantage of a high-throughput genotyping system that uses mass spectrometry measurements, the researchers were able to detect mutated mitochondrial DNA at high sensitivity. In each positive case, DNA cloning and sequencing were used to confirm the findings. By looking at differences in tissue samples from mother and child, the researchers were also able to estimate the rate at which new DNA mutations had arisen in the population. The incidence of new mutations was close to 100 for every 100, 000 live births. Dr. Samuels commented: “These new clinical measurements have given direct evidence for the widespread incidence of pathogenic mitochondrial DNA mutations in the human population. These findings emphasize the pressing need to develop effective ways to interrupt the transmission of these mutations to the next generation.” Source: Virginia Tech |
| Isn’t this just cool? [Mailund on the Internet] Posted: 12 Aug 2008 12:43 PM CDT Take a look at this post from Genetic Future. It concerns the genetic population structure of Europe, and it seems that the genotypes can be used to pretty accurately predict the geography. Of course I expected a lot of population structure when considering all of Europe, but I am a bit surprised that there is this much structure between, e.g., Swedes and Danes. I haven’t read the paper refered to yet — I will print it when I get to the office tomorrow — but this is pretty cool. What does this mean for association mapping, by the way. Is the HapMap CEU sample based tagSNPs equally good for all Europeans, or what? |
| New office [Mailund on the Internet] Posted: 12 Aug 2008 09:38 AM CDT
From my window I can see the old BiRC building. I don’t know what it will be used for in the future, assuming they do not just tear it down. It is an old building and it is not in a particularly good shape any more. Sorry about the picture quality, by the way. The pictures were taken by my cell phone and the quality is lousy. |
| Scientific Events in Second Life? [ScienceRoll] Posted: 12 Aug 2008 09:38 AM CDT What can you do if you would like to know what kind of scientific and health events you should attend in Second Life? Here are some useful links:
 |
| Posted: 12 Aug 2008 08:57 AM CDT |
| Genome Alberta Joins the DNA Network [Genome Blog] Posted: 12 Aug 2008 08:45 AM CDT We're pleased to note that Genome Alberta has been added to the DNA Network of blogs. It isn't easy being unique on the Internet but the DNA Network has certainly carved out a unique niche as a collection of more than 50 blogs covering the fields of genetics, genomics, and health. The network was co-founded by Dr. Hsien-Hsien Lei and Ricardo Vidal and has a varied range including Ricardo's 'My Biotech Life', 'Omics! Omics!', 'Reporter Gene', and Hsien's 'Eye on DNA'. Hsien recently moved from the U.K. to Singapore and Ricardo landed in the U.S from Portugal so you're assured of some international flavour if you follow the feed. We'll be adding the DNA feed to our main blog page so anytime you come to our pages you'll be able to see what's up on the DNA Network at the same time. That's a lot of blog reading to do but we'll do our best to make sure that the Genome Alberta pages are a kind on one-stop-blog-shop for you. You can also find the DNA Network on Facebook and of course don't forget to give a virtual gene on Facebook using the Genomics Application and check out Genome Alberta's group on Facebook. You may have noticed we've also decided to have a little fun with the world of genomics with Tammy Grady's Biopunk blog on these pages. Rightly or wrongly genetics has a strong presence in online gaming, home gaming systems, fiction, and fantasy. Tammy will be writing about that world and if you suspend your science mind for a few minutes you might find there is a whole world of genomics you never knew existed ! And last but not least we're also pleased to be added to BIO's own blog pages and be included as a guest blogger. The pages are fairly new and really first saw the light of day at BIO International in San Diego, and I'm looking forward so seeing how it develops between now and BIO Atlanta next year. |
| Climate Changed Debunked [Sciencebase Science Blog] Posted: 12 Aug 2008 07:00 AM CDT
If you’re in the UK, or have figured out the BBC iPlayer hack to let you use that tool outside the UK, you may have seen the recent global-warming-coming-oil-crisis-we’re-all-doomed drama Burn Up. You probably also heard about a little fella called Al Gore and his inconvenient movie and the Channel 4 documentary that attempted to shred it, perhaps a little conveniently ignoring some key facts as it did so. Meanwhile, power companies report massive profits and price rises for gas and electricity. They simultaneously pump up prices from well to wheel as the oil price bounces like a proverbial vulcanised rubber ball and everyone is looking to save gas. Anyway, I thought I’d post a shortlist of concerns expressed by more than a few people about the call to arms regarding climate change, seeing as I’ve published several items about alternative energy sources recently. I still stand by the didact: waste not, want not. It’s important that we cut pollution and it’s important that we reduce the amount of energy we waste. But, given that even the IPCC says there’s a 10% chance that climate change is not anthropogenic, maybe we should be looking at what we are planning for the world, especially in light of madcap schemes like adding lime to the oceans, before it’s too late. So, how does one answer the climate skeptics when faced with the following alleged myths?
|
| A Microcosm for Biology [Bitesize Bio] Posted: 12 Aug 2008 05:11 AM CDT I finally got around to purchasing and reading a copy of Carl Zimmer’s Microcosm: E.coli and the New Science of Life, and I have to chastise myself for not reading it sooner. In Microcosm, Zimmer has eloquently condensed a century of scientific study surrounding Eschericia coli into an accurate and flowing story readable by anyone with even just a modest understanding of biology.
And of course no book on E.coli would be complete without re-tracing its role in molecular biology. A long series of discoveries, made with E.coli as the experimental system, have elucidated the mechanisms of DNA replication and transcription, regulation of gene expression, and basic metabolism. Even genetic engineering techniques were pioneered in E.coli, which Zimmer describes in the chapter on “Playing Nature” - a nice twist on the old saying “Playing God,” that is actually more appropriate. Then there’s the story of E.coli’s vast evolutionary potential - from antibiotic resistence to immune evasion tactics, the simple and rapid replication cycle of bacteria have enabled natural selection, ecological niches, and population divergence to be studied over the course of tens of thousands of generations. In the process of his story, Zimmer explains how the bacterial genome is more of a palimpsest rather than an instruction manual - a book that’s been written and re-written many, many times. It’s that palimpsest that serves as both a history book of how it has been modified from its ancestors, but also as an example of “Open Source” text available to modification by its descendents and accessible to horizontal gene transfer. Lastly and in addition to references to Monod’s quote, the role of one oft-mentioned scientist is a microcosm of E.coli’s progress in biology - Joshua Lederberg. Zimmer writes of Lederberg in 1957 (p.193):
|
| Interesting News in the World of Genetic Genealogy [The Genetic Genealogist] Posted: 12 Aug 2008 02:00 AM CDT Technology Review, an MIT publication, has an article entitled “Genealogy Gets More Precise: Rapidly growing databases enable a more complete picture of one’s ancestry.“ The article, which is relatively balanced, discusses some of the benefits and challenges associated with genetic genealogy testing. Also check out the article and video “Mapping Out a Nascent Market” at boston.com, which is directed towards personal genetic companies such as deCODEme, 23andMe, Navigenics, and Knome. And lastly, scientists have sequenced and recreated the Neanderthal mtDNA genome. For more information see john hawks weblog, Genetic Archaeology, Genea-Musings (with a humorous twist), Anthropology.net, and The Spittoon. The original article is in Cell. Turns out there are roughly 206 differences between the CRS (the Cambridge Reference Sequence, the mtDNA to which all human mtDNA is compared) and Neanderthal mtDNA; 195 transitions and 11 transversions. |
| Local recent selection in humans [Yann Klimentidis' Weblog] Posted: 11 Aug 2008 08:34 PM CDT In this paper the authors use a combined Fst and haplotype block length approach to finding genes/areas that have been under strong recent selection in different populations: about 24 individuals each, of African American, European American, and Han Chinese descent, with 1.6 milion SNPs typed in each. After identifying haplotype blocks, they compared the average Fst for all SNPs in each block between populations. Genes involved in skin pigmentation for which they find evidence of positive selection:RAB27A, DCT, EGFR, ATRN, MATP. They also find that many of the areas under selection are in non-genic locations. They mention the example of the lactase gene is an example of the functional variant being in a regulatory region -- see this recent, interesting article in Science regarding the issue of the importance of genic vs. nongenic variation in evolutionary change (which this picture refers to).  Another interesting finding which they discuss is the small amount of overlap between loci that they find and the loci that other groups have found to be under recent positive selection. Identification of local selective sweeps in human populations since the exodus from Africa Åsa Johansson and Ulf Gyllensten, Hereditas Volume 145, Issue 3, Pages 126-1372008 ABSTRACT: Selection on the human genome has been studied using comparative genomics and SNP architecture in the lineage leading to modern humans. In connection with the African exodus and colonization of other continents, human populations have adapted to a range of different environmental conditions. Using a new method that jointly analyses haplotype block length and allele frequency variation (FST) within and between populations, we have identified chromosomal regions that are candidates for having been affected by local selection. Based on 1.6 million SNPs typed in 71 individuals of African American, European American and Han Chinese descent, we have identified a number of genes and non-coding regions that are candidates for having been subjected to local positive selection during the last 100 000 years. Among these genes are those involved in skin pigmentation (SLC24A5) and diet adaptation (LCT). The list of genes implicated in these local selective sweeps overlap partly with those implicated in other studies of human populations using other methods, but show little overlap with those postulated to have been under selection in the 5–7 myr since the divergence of the ancestors of human and chimpanzee. Our analysis provides focal points in the genome for detailed studies of evolutionary events that have shaped human populations as they explored different regions of the world. |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |






 So, how’s that for a blog post title? Catchier even than last Saturday’s
So, how’s that for a blog post title? Catchier even than last Saturday’s 

1 comment:
Amazing science! Thanks for posting the dog information.
Post a Comment