The DNA Network |
| Eye Candy - Nuclear Blebs [The Daily Transcript] Posted: 05 Aug 2008 07:37 PM CDT
| |
| I pick myself up, dust myself off, and I [Discovering Biology in a Digital World] Posted: 05 Aug 2008 02:11 PM CDT start all over again. MDRNA Inc., a Puget Sound area company formerly known as Nastech, announced on Monday that they'd be laying off 23 people including their president and chief business officer. This might not sound like a lot, but according to Joseph Tartakoff, from the Seattle PI, this brings the total number of layoffs up to 145 since November. These events present a challenge to those of us who teach in biotech programs or biotech-related fields. Nastech, the predecessor to MDRNA had been around for over 20 years. Who would expect a 20+ year company to shed three quarters of it's employees in a few short months? Read the rest of this post... | Read the comments on this post... | |
| Blogs by scientists. [Genomicron] Posted: 05 Aug 2008 11:49 AM CDT In case you aren't reading them yet, take a minute to check out these relatively new blogs by scientists: BdellaNea A blog about leeches -- Mark Siddall (American Museum of Natural History) Evolutionary Novelties A blog about evolution, with a soft spot for ostracod(e)s and eyes -- Todd Oakley (University of California Santa Barbara) The Rough Guide to Evolution A blog loosely accompanying a soon to be released book of the same name -- Mark Pallen (University of Birmingham) Chance and Necessity A blog about evo with a twist of devo -- Anonymous "Faculty member in the South". At least three of these bloggers are high end researchers, so do have a look. | |
| Religious diversity driven by disease prevalence [Yann Klimentidis' Weblog] Posted: 05 Aug 2008 10:22 AM CDT They invoke the optimal outbreeding hypothesis as to why out-group contact would be costly, but there is very scant evidence for this in humans. Then again, there's the paper I just posted about a few days ago that showed higher rates of obesity among multi-racial individuals, but I don't think that really counts in this context... Razib touches on some other potential examples here, ...as well on this paper. Assortative sociality, limited dispersal, infectious disease and the genesis of the global pattern of religion diversity Corey L. Fincher, Randy Thornhill Proceedings of the Royal Society, B First Cite Abstract: Why are religions far more numerous in the tropics compared with the temperate areas? We propose, as an answer, that more religions have emerged and are maintained in the tropics because, through localized coevolutionary races with hosts, infectious diseases select for three anticontagion behaviours: in-group assortative sociality; out-group avoidance; and limited dispersal. These behaviours, in turn, create intergroup boundaries that effectively fractionate, isolate and diversify an original culture leading to the genesis of two or more groups from one. Religion is one aspect of a group's culture that undergoes this process. If this argument is correct then, across the globe, religion diversity should correlate positively with infectious disease diversity, reflecting an evolutionary history of antagonistic coevolution between parasites and hosts and subsequent religion genesis. We present evidence that supports this model: for a global sample of traditional societies, societal range size is reduced in areas with more pathogens compared with areas with few pathogens, and in contemporary countries religion diversity is positively related to two measures of parasite stress. | |
| Posted: 05 Aug 2008 09:30 AM CDT This the second part of three part case study where we see what happens when high school students clone and sequence genomic plant DNA. In this part, we do a bit of forensics to see how well their sequencing worked and to see if we can anything that could help them improve their results the next time they sequence. How well did the sequencing work? | |
| Health Highlights - August 5th, 2008 [Highlight HEALTH] Posted: 05 Aug 2008 07:57 AM CDT
.gif) Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership! Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership!This article was published on Highlight HEALTH. Other Articles You May Like | |
| Science as Culture [Bitesize Bio] Posted: 05 Aug 2008 05:52 AM CDT Lawrence Krauss, one of the best popularizers of science since Carl Sagan, has another article in last week’s Science: Celebrating Science as Culture. In it, he reviews the World Science Festival that took place in New York City from May 28 to June 1, 2008. In the article itself, Krauss provides the big picture:
The culture of science has been aptly juxtaposed to that of the humanities since the 1959 lecture by C.P. Snow on The Two Cultures. And with today’s culture wars between the religion and secularism, the developing culture of science is contrasted even more starkly.
But it’s strange to think of science as a culture, especially for the scientists that are its lifeblood. We could care less about the cultural ramifications of our work; our attention is usually devoted to the work itself. Not that it is unimportant however. The sciences and the pragmatic realities that they study are universal, traditional religions and cultures are instead parochial, establishing the intrinsic tendency of ethnocentrism and intolerance that we blithely attribute to “Human nature.” But I digress. Science has become a culture, in fact. The Oxford Dictionary describes “Culture” as “Learned behavior which is socially transmitted, such as customs, belief, morals, technology, and art; everything in society which is socially, rather than biologically transmitted.” Science, from graduate school onward, definitely fits into that description. From your thesis to the daily lab-work routine to the sacredness of lab notebooks to taboos against fabricating data, science is culture. We often are as passionately engaged in our work as anyone else. It even feels cool to be a geek, when one finds him or herself in “geek culture.” Image credit: Phillip Torrone | |
| Are Disease-Causing mtDNA Mutations Common? [The Genetic Genealogist] Posted: 05 Aug 2008 02:00 AM CDT A new study in the American Journal of Human Genetics (available here) examined the frequency of ten (potentially) pathogenic mitochondrial point mutations in 3168 neonatal cord blood samples. Of these samples, a total of 15 (or 1 in 200) harbored one or more of the mutations. Interestingly, the mtDNA of 12 of the 15 samples were heteroplasmic, meaning that their cells harbored both mutated and non-mutated mtDNA genomes. Figure 1 from the paper, above, shows the percentage of mutated mtDNA in each of the 15 samples with mutations, from nearly 0% to the 100% in the three homoplasmic samples. The abstract:
HT: Dienekes’ Anthropology Blog | |
| Charles Darwin: Genius [HENRY » genetics] Posted: 05 Aug 2008 12:43 AM CDT | |
| Posted: 05 Aug 2008 12:15 AM CDT Henn et al. in P.N.A.S. present Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa (doi):
| |
| ETech CFP calls out personalized medicine, synbio [business|bytes|genes|molecules] Posted: 04 Aug 2008 11:31 PM CDT Brady Forrest has announced the Call for Papers for ETech. Two of the topics should be of interest to bbgm readers Personalized Healthcare: Medical technology is something that almost everyone comes to rely on, whether it's hopeful, preventive care in the form of Reseveratol, or a new limb. In no other area does the industrialized world have more of an advantage. What legal framework for personal genomics balances innovation and appropriate medical caution? How is medicine changing? How is healthcare changing across the world? Many resources are focused on anti-aging technology and drugs—is this the right direction? Synthetic Biology: We can't cover the reinvention of living without looking at the new definition of life. Synthetic biology, first pioneered in the 1970s, is becoming a factor in the development of new materials, medicines, environmental cleansing, and energy. How will this technology impact our lives? How can we be a part of it? What will bring it into the hands of the wider public? I hope that the online life science community can submit something. Would be nice to see some real life science geeks there. Plus Timo’s on the selection committee Of course, those are not just the only cool topics. | |
| What do you do when a creationist asks for your data? [adaptivecomplexity's column] Posted: 04 Aug 2008 11:20 PM CDT I apparently missed this little episode in June, when a creationist (with no scientific credentials) read a news piece on the recent work of Michigan State biologist Richard Lenski, and then wrote an obnoxious letter demanding Lenski's data. Lenski has had a long-running bacterial evolution project, and recently published a paper on the evolution of citrate-metabolizing bacteria in his lab. Lenksi has long been the focus of creationist attacks, because much of his in-the-lab evolutionary experiments strike right at the heart of the claims of the intelligent design movement. This latest issue has been well-covered in various science blogs, but this little scene raises an interesting question: are scientists obligated to share data or reagents with anyone who asks, scientist or not? | |
| Discussing publishing at ISMB [business|bytes|genes|molecules] Posted: 03 Aug 2008 12:46 PM CDT BioInform has a report (sub reqd, or get it from the Google cache) on a special session on scientific publishing organized my Scott Markel at ISMB. Since that is a topic of particular interest to bbgm and bbgm readers, thought I’d give it a read and find out if there were any new insights there. I won’t delve into all the specifics, but it is clear that there is a desire in the community to make scientific publication as machine readable as possible. As has been discussed many times here, scientific research is a treasure trove of information and the content contained within papers and other forms of publishing should become part of the data that we try to mine and correlate, and in the long term it’s not just the text, but as pointed out in the article images and other datasets as well. Mark Gerstein in particular brings up some points which make a ton of sense, and to a degree are somewhat obvious. That we still need to debate them is the part that makes me frown. Wouldn’t it be nice if we could take some of those ideas and thoughts for granted? For example
The part that people don’t seem to grasp, or at least it didn’t jump out to me is that for text-based documents, we have a database, one called the world wide web. Via DOIs and other identifiers, each paper is an addressable resource. Given the right structure and the right APIs it’s a data mashup waiting to happen. Or if you structure things the right way, and want access to a nice n-tuple data store, use something like Freebase or Talis as a backend platform But I agree with Matt Cockerill as well. We need better authoring tools, and authoring tools need standards for markup and structure. I am not sure where this comes from and how this will be implemented. Unfortunately, we all still write our papers in Word. The LaTeX crowd, or the HTML crowed would probably be happy providing some markup (it doesn’t have to be too heavy). I won’t even debate open access vs. closed access. In my mind it is no longer a debate. It is good to hear publishers talking about online journals, and about integrating other media formats into publications. |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |
 In contrast to the evidence-based approach to the world provided by science, where building knowledge and recognizing uncertainty are central, and where humanity is not divided by boundaries, the alternative foundations for culture fall flat on their faces.
In contrast to the evidence-based approach to the world provided by science, where building knowledge and recognizing uncertainty are central, and where humanity is not divided by boundaries, the alternative foundations for culture fall flat on their faces. 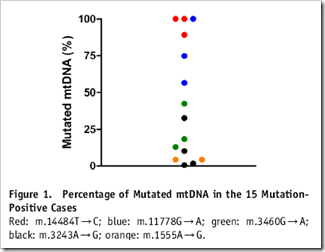 Genetic genealogy has the potential to reveal information about your health (for example,
Genetic genealogy has the potential to reveal information about your health (for example,  on haplogroup J implicates the background mtDNA haplotype in mutagenesis. These findings emphasize the importance of developing new approaches to prevent transmission.”
on haplogroup J implicates the background mtDNA haplotype in mutagenesis. These findings emphasize the importance of developing new approaches to prevent transmission.”


No comments:
Post a Comment