The DNA Network |
| miRNA Pioneers take the Lasker [The Daily Transcript] Posted: 15 Sep 2008 09:31 PM CDT I've been neglecting science lately but I just wanted to point out that Victor Ambros, Gary Ruvkun and David Baulcombe just won the Lasker Award in Basic Sciences for their work on miRNAs, small non-coding RNAs that are encoded in the genome. These 19-23 nucleotide long RNAs regulate the stability, localization and translation of mRNAs and have been implicated in almost every biological process from stem cells, to cancer to development. Ambrose and Ruvkin are well known for their work on miRNAs and worm development, while the lesser known Baulcombe made similar discoveries in plants. These findings (along with RNAi in general) have led to one of the most important conceptual advances in biology within the past few decades. |
| Let's talk about the facts this election - Part IX - Offshore Drilling [The Daily Transcript] Posted: 15 Sep 2008 06:40 PM CDT Here we are going to look at the best available figures for offshore drilling, specifically the areas that are currently off-limits. That's what the fight is about. First, how much oil we consume and how much we "produce":
The bottom line is that we consume a heck-of-a-lot more, close to twice what we currently produce. And how much more could we get "offshore" from areas that are currently off-limits? Well if you comb all the literature out there, the simple answer is that we don't really know. Here's the closest that I've been able to get from an article in Scientific American: The Minerals Management Service (MMS), part of the U.S. Department of the Interior responsible for leasing tracts to oil and gas companies and collecting the royalties on them ... has estimated that there are around 18 billion barrels in the underwater areas now off-limits to drilling. That's significantly less than in oil fields open for business in the Gulf of Mexico, coastal Alaska and off the coast of southern California, where there are 10.1 billion barrels of known oil reserves as well as an estimated 85.9 billion more. So the whole fight is for 18 billion barrels (as far as we know). To place this figure into context, the current estimated reserves for the US (excluding all offshore) is about 22 billion barrels. Add 96 billion barrels from the current offshore drilling areas (total now up to 118 billion barrels) and those 18 billion barrels don't seem like so much. Let's look at the world's oil reserves and consumption rates and compare all these numbers:
Note that by 2030, oil consumption is estimated to go way up due in large part to China and India. You'll also notice that the amount of oil in these offshore off-limit areas are likely to be negligible in the long run. Fully exploring the areas that we currently drill will likely produce far more oil. Opening the off-limit areas will not affect the world supply and will certainly have little effect on the price of gas. But it will line the pockets of anyone who can drill. Remember the price of gas is largely dictated by world supply and world demand - the notion that the US is an insulated market is hogwash. So why would John McCain and the GOP support such a measure? Sources: Energy Supply page on the Global Education Project, The Energy Information Administration, The Minerals Management Service, The CIA Factbook. Read the comments on this post... |
| Admixed EUR-EAS population [Yann Klimentidis' Weblog] Posted: 15 Sep 2008 05:29 PM CDT  Apparently, an admixed population where the admixture event happened 120 generations ago can be useful for admixture mapping, and for studying diseases that differ in prevalence between Europeans and East Asians. For some reason, my university is not giving me full text access which is frustrating. Apparently, an admixed population where the admixture event happened 120 generations ago can be useful for admixture mapping, and for studying diseases that differ in prevalence between Europeans and East Asians. For some reason, my university is not giving me full text access which is frustrating.A Genome-wide Analysis of Admixture in Uyghurs and a High-Density Admixture Map for Disease-Gene Discovery.
Abstract: Following up on our previous study, we conducted a genome-wide analysis of admixture for two Uyghur population samples (HGDP-UG and PanAsia-UG), collected from the northern and southern regions of Xinjiang in China, respectively. Both HGDP-UG and PanAsia-UG showed a substantial admixture of East-Asian (EAS) and European (EUR) ancestries, with an empirical estimation of ancestry contribution of 53:47 (EAS:EUR) and 48:52 for HGDP-UG and PanAsia-UG, respectively. The effective admixture time under a model with a single pulse of admixture was estimated as 110 generations and 129 generations, or admixture events occurred about 2200 and 2580 years ago for HGDP-UG and PanAsia-UG, respectively, assuming an average of 20 yr per generation. Despite Uyghurs' earlier history compared to other admixture populations, admixture mapping, holds promise for this population, because of its large size and its mixture of ancestry from different continents. We screened multiple databases and identified a genome-wide single-nucleotide polymorphism panel that can distinguish EAS and EUR ancestry of chromosomal segments in Uyghurs. The panel contains 8150 ancestry-informative markers (AIMs) showing large frequency differences between EAS and EUR populations (F(ST) greater than 0.25, mean F(ST) = 0.43) but small frequency differences (7999 AIMs validated) within both populations (F(ST) less than 0.05, mean F(ST) less than 0.01). We evaluated the effectiveness of this admixture map for localizing disease genes in two Uyghur populations. To our knowledge, our map constitutes the first practical resource for admixture mapping in Uyghurs, and it will enable studies of diseases showing differences in genetic risk between EUR and EAS populations. |
| Vaccine against HER2-positive breast cancer offers complete protection in lab [Think Gene] Posted: 15 Sep 2008 05:02 PM CDT Josh: I’ve been waiting for a study such as this to come out. We will never be able to successfully fight off cancer with drugs alone. Cancer is a normal part of life; many cells in each of our bodies are cancerous, but our immune systems successfully destroy them. When people are actually diagnosed with cancer, the immune system for some reason stopped recognizing the cells as cancerous. This technique basically fixes that, allowing the body to fight it off. I’m not surprised in the slightest that all traces of cancer were gone. The next step is to try this in humans, probably just testing the immune response first, then introducing it to some cancer patients. I see no reason why this can’t be applied to all other types of cancer, as long as they have some type of unique, recognizable receptor or surface marker. Perhaps I’m just an optimist, but I’m holding out hope that this is the beginning of the end of the search for a cure to cancer. Researchers at Wayne State University have tested a breast cancer vaccine they say completely eliminated HER2-positive tumors in mice - even cancers resistant to current anti-HER2 therapy - without any toxicity. The study, reported in the September 15 issue of Cancer Research, a journal of the American Association for Cancer Research, suggests the vaccine could treat women with HER2-positive, treatment-resistant cancer or help prevent cancer recurrence. The researchers also say it might potentially be used in cancer-free women to prevent initial development of these tumors. HER2 receptors promote normal cell growth, and are found in low amounts on normal breast cells. But HER2-positive breast cells can contain many more receptors than is typical, promoting a particularly aggressive type of tumor that affects 20 to 30 percent of all breast cancer patients. Therapies such as trastuzumab and lapatinib, designed to latch on to these receptors and destroy them, are a mainstay of treatment for this cancer, but a significant proportion of patients develop a resistance to them or cancer metastasis that is hard to treat. This treatment relied on activated, own-immunity to wipe out the cancer, says the study’s lead investigator, Wei-Zen Wei, Ph.D., a professor of immunology and microbiology at the Karmanos Cancer Institute. “The immune response against HER2-positive receptors we saw in this study is powerful, and works even in tumors that are resistant to current therapies,” she said. “The vaccine could potentially eliminate the need to even use these therapies.” The vaccine consists of “naked” DNA – genes that produce the HER2 receptor – as well as an immune stimulant. Both are housed within an inert bacterial plasmid. In this study, the researchers used pulses of electricity to deliver the injected vaccine into leg muscles in mice, where the gene produced a huge quantity of HER2 receptors that activated both antibodies and killer T cells. “While HER2 receptors are not usually seen by the immune system when they are expressed at low level on the surface of normal cells, a sudden flood of receptors alerts the body to an invasion that needs to be eliminated,” Wei said. “During that process, the immune system learns to attack cancer cells that display large numbers of these receptors.” They also used an agent that, for a while, suppressed the activity of regulatory T cells, which normally keeps the immune system from over-reacting. In the absence of regulatory T cells, the immune system responded much more strongly to the vaccine. Then, when the researchers implanted HER2-positive breast tumors in the animals, the cancer was eradicated. “Both tumor cells that respond to current targeted therapies and those that are resistant to these treatments were eradicated,” Wei said. “This may be an answer for women with these tumors who become resistant to the current therapies.” Wei’s lab is the first to develop HER2 DNA vaccines, and this is the second such vaccine Wei and her colleagues have tested more extensively. The first, described in a study in 1999, formed the model of a vaccine now being tested by a major Pharmaceutical company in early phase clinical trials in the U.S. and in Europe in women with HER2-positive breast cancer. In order to ensure complete safety, Wei says the current test vaccine uses HER2 genes that are altered so that they cannot be oncogenic. The receptors produced do not contain an “intracellular domain” – the part of the receptor that is located just below the cell surface and transmits growth signals to the nucleus. The first vaccine was also safe, she says, but contained a little more of the native HER2 receptor structure. “With this vaccine, I am quite certain the receptor is functionally dead,” she said. “The greatest power of vaccination is protection against initial cancer development, and that is our ultimate goal with this treatment,” Wei said. Source: American Association for Cancer Research |
| Lake arrowhead notes [The Tree of Life] Posted: 15 Sep 2008 01:42 PM CDT Well I gave my talk Seemed to go ok except getting cutoff early because the chair ignored George Weinstock is now speaking using my laptop so I am trying to He said one key thing I left out ... Big scale microbial sequencing More later ------------------------------------------------- George Weinstock gave a good overview of the "Human Microbiome Project" which is a NIH Roadmap initiative to catalogue the genomic content of the microbes associated with humans. He described some of the big picture of why do the project, of the different fundingin initiatives being done through NIH and he gave some detail on the "jumpstart" project going on at the big genome centers right now. He outlined how the current plan is to select a few hundred people and to survey their mcirobiomes from multiple sites using rRNA PCR and possibly metagenomics. In addition, he described how there is also an effort to sequence 100s if not a 1000 genomes of cultured organisms that have been isolated from human environments. He did say one thing I disagreed with which is that he thinks it is somewhat reasonable to treat the environment that microbes live in in essence as a big bag of genes. In other words, if you sequence from a community, he implied that one can focus just on the genes and their functions and not the organisms that they come from. On this I disagree (and pointed this out after the next talk). But overall George gave anice overview of the project and its goals. Eric Womack gave a good talk about viral metagenomics work he has been doing. He pointed out that a lot of the viral world is "unknown" but that does not mean it is unimportant. And this is consistent with what I and George Weinstock said which is that we need more genome data from viral isolates. Eric presented some very useful results on the challenges of using short read sequence data in metagenomics and he referenced a few papers on this. He also referred to a cool viral genome survey project that I was not aware of by Hatfull which involved undergraduates in sequencing and analyzing the genomes of phage that infect Mycobacterium smegmatis. Jim Bristow on Biofuels. He is now giving a summary of some of the JGI work on the genomics of cellulolytic organisms and processes. He is focusing on the termite gut community and had some good one liners about this (e.g., he said many people want to kill termites but not JGI. They are our friends; he also said "it takes a village to sequence a termite gut"). |
| Simultaneous analysis of all SNPs in a genome-wide association study [Mailund on the Internet] Posted: 15 Sep 2008 12:10 PM CDT In our association mapping journal club a few weeks back, we discussed this paper (I just never got around to writing down my thoughts on it until now):
I already heard about the method when I was visiting Imperial College to give a seminar last year, so I am happy that I can finally talk about it. It is a pretty neat idea. Regression analysis in association mappingIf you want to figure out which parameters are important for predicting some property, a good old statistical approach is regression analysis. For a binary property, such as case or control in an association study, you could use logistic regression, but in general you construct some linear function of your parameters and transform them into the “property space” through a link function. This setup gives you a “model” and depending on the link and the setup you have different ways of interpreting this as a statistical model with a corresponding likelihood function. The coefficients in the linear combination of parameters are the parameters in the model, and you typically maximize the likelihood with respect to them to get your estimate for them. In some cases you can directly interpret the parameters, but more often than not you are only interested in knowing whether there is strong evidence in the data that they should be non-zero, i.e. that the parameter in question actually has an effect on the property. In an association study, you would use your SNPs as your parameters and you consider those SNPs with a non-zero coefficient associated with the disease. Of course, it is never as simple as that. Two things complicate matters: your best estimate of a coefficient will never actually be zero, so you want to test if they are significantly different from zero. Another problem is that you have many more parameters (SNPs) than you have outcomes (individuals), so you will overfit from hell. Strong “zero” priorsWhat they do in this paper is both simple and very clever. They consider the problem in a Bayesian setting and put strong priors on the coefficients, that will tend to keep them at zero unless the signal in the data is strong enough to pull them away from there. They then test for association by testing if the mode of the posteriors for these parameters have moved away from zero. A very nice consequence of this is that you can analyse the entire data at the same time, rather than testing markers individually, which means that if several markers are in LD with a causal marker, you will tend to only pick one of them and recognize that the signal in the others is essentially the same signal. It also seems quite computationally feasible. A few hours on a desktop computer to analyse a GWA data set. Clive J. Hoggart, John C. Whittaker, Maria De Iorio, David J. Balding, Peter M. Visscher (2008). Simultaneous Analysis of All SNPs in Genome-Wide and Re-Sequencing Association Studies PLoS Genetics, 4 (7) DOI: 10.1371/journal.pgen.1000130 |
| My Son, The Genetic Epidemiologist [Eye on DNA] Posted: 15 Sep 2008 11:18 AM CDT My six-year-old’s reading and mark-up (in purple) of a paper in Nature Genetics authored by my friend, Dr. Linda Kao. Press Release - New gene variant identified for nondiabetic end stage renal disease in African-Americans
|
| iPS cell lines developed for disease research and drug discovery [Mary Meets Dolly] Posted: 15 Sep 2008 10:36 AM CDT
I have read repeatedly that using human embryos in this way is not only important but necessary. Not so much. Researchers have created 10 pluripotent stem cell lines with diseases ranging from Parkinsons to diabetes. These lines did not come from embryos, but from patient cells that have been reprogramed or "induced" back to pluripotency. They call these iPS cell lines. From Cell:
These iPS lines are now going to be used for research into these diseases. The ethical alternatives keep on coming. |
| New genetic variant discovered, associated with bladder cancer [Mailund on the Internet] Posted: 15 Sep 2008 10:20 AM CDT I just saw this press release: deCODE and Radboud University Discover Common Variants in the Human Genome Conferring Risk of Bladder Cancer We, here at BiRC, actually collaborate with both deCODE and Radboud Uni in the EU PolyGene project. The bladder cancer analysis is not part of PolyGene, but through the collaboration we have access to it, and we have just started analysing it. We weren’t in on the initial analysis, though, so we are not part of this discovery. We only get access to data after they have already mined what they can find themselves. A bit annoying, but perfectly reasonable. Our contribution to the collaboration is methods development, and anything they can find with the methods they already have, they do not really need us for. Still, it would have been nice to be in on the analysis from the beginning. Whenever we get our hands on the data, we always get excited about hits only to discover that they are already submitted for publication. Anyway, nice to see that they get something out of the data. |
| Don’t try this at home…? [genomeboy.com] Posted: 15 Sep 2008 10:09 AM CDT Mackenzie Cowell (left) and Jason Bobe are trying to create simple, at-home methods for doing sophisticated biology. (Dina Rudick/Globe Staff)
One wonders if maybe we couldn’t use a few more lunatics. |
| I really shouldn’t work with computes… [Mailund on the Internet] Posted: 15 Sep 2008 10:03 AM CDT Only a few months ago, my Mac died on me. Today my Linux box did. I have no idea what is wrong with it, but I can no longer boot it up. Both machines are less than a year old, so I think it is a bit early for them to die on me… |
| How do you really feel, Dr. Wakeley? [evolgen] Posted: 15 Sep 2008 08:00 AM CDT I'm currently working my way through John Wakeley's book on Coalescent Theory. (The website has a few pre-publication chapters if you want to take a peek.) In his introductory chapter, Wakeley introduces the concept of gene genealogies. He's careful to point out that, unlike the phylogenies we construct using inter-specific data, we don't actually use intra-specific gene genealogies to infer the relationships of the sequences we've sampled:
Basically, Wakeley is calling out two high-profile researchers for their role in blurring the line between gene genealogies and phylogenetics: Alan Templeton and John Avise. Avise gets credited for creating phylogeography, while Templeton is responsible for nested clade analysis. Both of these approaches have classically ignored the role stochastic processes play in shaping intra-specific genetic variation. In doing so, practitioners of phylogeography often invoke just-so-stories to explain the demography history of their species of interest. Read the rest of this post... | Read the comments on this post... |
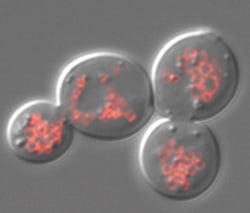
| Evolution of Lager Yeasts [Bitesize Bio] Posted: 15 Sep 2008 05:18 AM CDT For something a bit more on the fun side, at least if you enjoy a pint of beer now and then - a genomic-based study has reconstructed the origins by hybridization of the lager yeast Saccharomyces pastorianus, published in the journal Genome Research [Press release].
But genetically, it should not be surprising that these yeasts separate into distinct groups of strains, traceable to single events in brewing history. This study, which used array comparative genomic hybridization (aCGH), traced one such lineage. As the study showed though, S. pastorianus had two origins:
Maybe S. pastorianus should be re-classified into two yeast species, based on this data. I wonder whether these separate hybridization events are responsible for distinct types of lagers, and what those might be. Hat tip: Fungal Genomes |
| Democratization? Or Capitalization? Take yer pick [The Gene Sherpa: Personalized Medicine and You] Posted: 15 Sep 2008 03:48 AM CDT |
| New calcium reporter for two-photon imaging in vivo [Reportergene] Posted: 15 Sep 2008 03:17 AM CDT Marko Mank and colleagues started filling this gap by means of mutagenesis on the TnC calcium biosensor. These efforts increased overall signal strength and sensitivity in the regime of physiologically relevant calcium concentrations leading to a new biosensor, the TN-XXL, that is functional in vivo in flies and mice and, according to the authors, can be used to obtain tuning curves of neurons in visual cortex using in vivo two-photon imaging. As a perspective, the new reporter will be useful to get more insigths about calcium role into plasticity and degeneration. Marco Mank, Alexandre Ferrão Santos, Stephan Direnberger, Thomas D Mrsic-Flogel, Sonja B Hofer, Valentin Stein, Thomas Hendel, Dierk F Reiff, Christiaan Levelt, Alexander Borst, Tobias Bonhoeffer, Mark Hübener, Oliver Griesbeck (2008). A genetically encoded calcium indicator for chronic in vivo two-photon imaging Nature Methods, 5 (9), 805-811 DOI: 10.1038/NMETH.1243 |
| Spray-on Condoms [Sciencebase Science Blog] Posted: 15 Sep 2008 02:40 AM CDT
This supposedly original idea of applying Latex in spray-on form looks like an April Fool’s joke. First off, it’s not a new idea, especially given the range of colours the inventor is working with. I have heard of several patent applications for similar approaches to contraception and safer sex over the years, they even get a mention in Ben Elton’s book This Other Eden. The idea is fatally flawed on several fronts. In the heat of passion, I suspect that producing a laboratory-standard uniform layer with no weak points will be impossible and therefore make the device ineffective. However, entanglement of the material with pubic hair would also be a serious issue at the time of desheathing. It’s bad enough removing a band-aid from a grazed knee, but this has the potential to cause much worse pain. I assume that the process will be safety tested before it is made commercially available, but there are certain characteristics of an aerosol spray that could not be avoided. Primarily, the sprayer and teh sprayee are liable to be breathing more heavily than usual, to have slightly raised blood pressure, and perhaps be open mouthed. The last thing you would want to be near in such circumstances is close to airborne Latex - think potential inhalation, anaphylactic shock and risk of death. Such a spray would almost certainly be designed for external use only, and yet the organ destined to be coated not only has delicate surface tissues, but an aperture through which particles and carrier solvent might enter. Penile contact dermatitis anyone? Didn’t think so! And, speaking of solvents, presumably there will be a carrier solvent in which the Latex will be suspended and transported from spraycan to the surface to be coated. The phrase latent heat of evaporation comes to mind and its attendant rapid chilling effect, so there is also potential for frost-bite or at best stinging pain. One more issue. As you can see from the photo of the inventor creating a condom-shaped mess, the process of applying a spray-on condom does not look like a particularly neat and tidy one. If you have ever had difficulty explaining lipstick on your collar, then you will definitely struggle to explain a lurid yellow smear of rubber on your underwear. It almost makes abstinence seem like a viable option. |
| R.I.P. David Foster Wallace [adaptivecomplexity's column] Posted: 14 Sep 2008 11:13 AM CDT David Foster Wallace committed suicide last Friday. The NY Times has an appraisal here. No more novels with inimitable passages like this: |
| Are the only good US Schools in the East? [adaptivecomplexity's column] Posted: 14 Sep 2008 10:29 AM CDT Today's Sunday Times reflects on the Western state of mind. The piece resonated with me, having grown up in an Eastern Ivy League town, but also having lived for seven years in three different Western states. Vice Presidential candidate Sarah Palin's University of Idaho credentials come up (and no, this post is not about politics or Sarah Palin!):
The University of Idaho is a bad choice to illustrate this point, |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |





 As a kind of follow-up to my
As a kind of follow-up to my
No comments:
Post a Comment