The DNA Network |
| Dinosaur: it’s what’s for dinner [genomeboy.com] Posted: 23 Sep 2008 08:02 PM CDT Munger, who writes the Progressive Alaska blog, told me Palin is not just a creationist, but a “young Earth” creationist who believes that man and dinosaurs once shared the planet, and that the world will end in her lifetime. | ||
| Posted: 23 Sep 2008 06:28 PM CDT  We discussed at lunch if you could survive, drinking your own urine. It was lunch so drinking other people's urine wasn't discussed as it seemed unappetizing. In any case I can't really figure out how long you could repeatedly drink your own urine to stave off dehydration. However you could distill your urine. Of course, I wouldn't bother with digging a pit ect. like the link suggests when you could just do it on your stove. In any case this reminded me that urine contains the element phosphorus, which has an interesting history. Check out a great synopsis on the history the discovery of phosphorus. Also there is an entire book out about phosphorus. Here is a review of The Shocking History of Phosphorus by John Emsley by the Naked Scientists. | ||
| What does privacy mean to you? Please complete this survey [The Tree of Life] Posted: 23 Sep 2008 06:21 PM CDT ThePrivacyPlace.Org Privacy Survey is Underway! Researchers at ThePrivacyPlace.Org are conducting an online survey about privacy policies and user values. The survey is supported by an NSF ITR grant (National Science Foundation Information Technology Research) and was first offered in 2002. We are offering the survey again in 2008 to reveal how user values have changed over the intervening years. The survey results will help organizations ensure their website privacy practices are aligned with current consumer values. The URL is: http://theprivacyplace.org/currentsurvey We need to attract several thousand respondents, and would be most appreciative if you would consider helping us get the word out about the survey, which takes about 5 to 10 minutes to complete. The results will be made available via our project website (http://www.theprivacyplace.org/). Prizes include $100 Amazon.com gift certificates sponsored by Intel Co. and IBM gifts On behalf of the research staff at ThePrivacyPlace.Org, thank you! | ||
| Open Access Under Attack [The Daily Transcript] Posted: 23 Sep 2008 05:09 PM CDT I'm back from Toronto. And now I'm just trying to keep up with all the crap I haven't dealt with in the last few days. Tomorrow we have an RNA Data club meeting (info here) and then I got this interesting email about some terrible legislation that might actually come to a vote tomorrow: On September 11, 2008, the Chairman of the House Judiciary Committee (Rep. John Conyers, D-MI) introduced a bill that would effectively reverse the NIH Public Access Policy, as well as make it impossible for other federal agencies to put similar policies into place. For more, read what Bora had to say. Read the comments on this post... | ||
| Gene-geography video [Mailund on the Internet] Posted: 23 Sep 2008 04:32 PM CDT A youtube video about this paper: You might also want to check out this paper. | ||
| Posted: 23 Sep 2008 04:10 PM CDT Every fall, we had to confront it. People would let their dogs run around on the field in the morning and by the time soccer practice started, the field would be full of deadly doo.
That's why I'm so exciting to see this new application for DNA testing. That's right. Kids will be able to play soccer without worry and dog owners will be held responsible for cleaning up after their pets. Read the rest of this post... | Read the comments on this post... | ||
| Google Founder, Parkinson’s Disease and Good Marketing? [ScienceRoll] Posted: 23 Sep 2008 03:39 PM CDT Sergey Brin, the co-founder of Google, wrote about his genetic predisposition for Parkinson’s Disease on his personal blog. Of course, he shared information from his 23andMe account, what else. His wife, Anne Wojcicki is the co-founder of 23andMe, a personalized genetic company.
Well, he didn’t analyze properly his genetic results as Steve Murphy, our gene sherpa and the clinical genetics fellow at Yale, pointed out some days ago. First, the gene Brin mentioned (LRRK2) is not the most important gene in the story of Parkinson’s and second, his risk cannot be 80%. According to Steve:
Kevin Fischer had some comments about it as well. Why was it a fantastic marketing trick?
Which means Brin is a good husband and knows how to promote her wife’s service efficiently.  | ||
| Any Happy Navigenics Customers Yet? [Week printf("%d",i++)!] [Think Gene] Posted: 23 Sep 2008 03:33 PM CDT We here at Think Gene are still waiting for a report of least one person who will vouch as a satisfied Navigenics customer. To qualify:
There is no prize for identifying oneself as this happy Navigenics customer because we feel that the joy of being a happy customer of Navigenics would be its own reward. A running tally of identified satisfied Navigenics customers will be kept below for your convenience:
Leave reports in the comments. | ||
| ScienCentral Channel: Genetic Genealogy [ScienceRoll] Posted: 23 Sep 2008 03:19 PM CDT I’ve been a subscriber of the ScienCentral channel in Youtube for a while and there are really great videos about important topics such as genealogy (Blaine will certainly like it).
 | ||
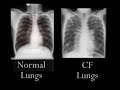
| Potential new treatment for cystic fibrosis? [ScienceRoll] Posted: 23 Sep 2008 03:12 PM CDT I just found an interesting article at the PHG Foundation about a new potential treatment for cystic fibrosis, a genetic condition affecting the exocrine (mucus) glands of the lungs, liver, pancreas, and intestines. It is caused by a mutation in the CFTR gene. The product of that gene is a a chloride ion channel that plays role in creating digestive juices and mucus. If there is no normal copy of the gene, the person will be affected by CF.
Here is a video describing the symptoms:  | ||
| Sub-Zero Bioprospecting: Tip Of The Iceberg [] Posted: 23 Sep 2008 02:45 PM CDT
It’s not just the tropics generating all the genetic diversity and commercially promising compounds. Nature responds to environmental extremes in all sorts of biotechnologically-leverageable (ouch. syllabic train-wreck.) ways. The Canadian Press reports on how DNA from arctic organisms can be applied: new antifreeze compounds in food products, new medical therapies for stroke victims, and more. International issues loom: “Canada may not be taking advantage of its extensive Arctic ecosystem. Of 31 known patents concerning products derived from Arctic organisms, two-thirds are held by U.S. companies. None are Canadian.” | ||
| Examining the Medical Blogosphere: An Online Survey of Medical Bloggers [ScienceRoll] Posted: 23 Sep 2008 01:34 PM CDT Some months ago, I was asked to take a survey focusing on the habits of medical bloggers. The research was conducted by Ivor Kovic et al. and they published the results in the Journal of Medical Internet Research (JMIR).
 | ||
| Advancing science through blogging [Mailund on the Internet] Posted: 23 Sep 2008 11:40 AM CDT There’s a paper in the latest issue of PLoS Biology about blogging: Advancing Science through Conversations: Bridging the Gap between Blogs and the Academy The topic is how blogs can be used as an outreach from scientists to the public. That sounds fair enough. There’s also a bit about how this can be formalised. I’m not sure I would appreciate that much. I like my blogs informal. For formal stuff, I’ll stick to journals. There’s a detailed comment on the paper at DrugMonkey. By and large I agree with the comments there… | ||
| Dick Eastman Interviews The Genetic Genealogist at FGS 2008 [The Genetic Genealogist] Posted: 23 Sep 2008 11:36 AM CDT On September 5th at the 2008 Federation of Genealogical Societies Conference in Philadelphia, Pennsylvania, I was interviewed by Dick Eastman. In the interview we discuss my blog, DNA testing in general, and my free ebook, “I Have the Results of My Genetic Genealogy Test, Now What?” (which is available for download in the sidebar of the blog). If the player doesn’t appear in the post, the interview is available here (http://rootstelevision.com/players/player_conferences.php?bctid=1811559654). It was a pleasure to meet and talk with Dick, and I hope you enjoy the interview. | ||
| You Know You Want a Yeast Biochemical Pathways Wall Poster [adaptivecomplexity's column] Posted: 23 Sep 2008 11:23 AM CDT Who wouldn't want a map of all the known yeast metabolic pathways on the wall? Download your copy here (PDF). The catch is you have to print it out yourself. | ||
| Do mosquitoes get the mumps?, part III [Discovering Biology in a Digital World] Posted: 23 Sep 2008 10:00 AM CDT Part III. Serendipity strikes when we Blink In which we find an unexpected result when we Blink while looking at the mumps polymerase. This is the third in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes. And yes, this is where the discovery happens. I. The back story from the genome record | ||
| Posted: 23 Sep 2008 09:00 AM CDT The National Girls Collaborative Project, as you might guess from the title, focuses on helping girls and engaging girls in science, technology, engineering, and math (aka "STEM"). photos used with permission from NGCP Read the rest of this post... | Read the comments on this post... | ||
| Profiling Python [Mailund on the Internet] Posted: 23 Sep 2008 08:33 AM CDT I’ve spent today writing some scripts to analyse a genome. I have about 2 giga bases of alignment, so it can be pretty slow to extract information from it. Especially since the cluster here at BiRC is in heavy use these days, so I cannot gain much from parallelising the analysis. Psyco and JIT compilingI had hoped that I could gain the speed I needed using Psyco. I’ve been lucky with that in the past, getting about x10 in speed. It is a JIT (just-in-time) compiler that actually compiles (generate machine code) the code rather than interpret it. With languages such as C++ you need to compile the code more or less manually before you can run it. That translate the high-level code into machine code that can actually run on the machine. In Java you also compile the high-level code, but there you compile it into something called byte code that cannot actually run on the machine. Instead the byte code can be interpreted in a Java Virtual Machine (JVM). This is how you get their “compile once, run everywhere”. This is actually also what Python does, you probably just haven’t noticed, because Python does it automatically. When it loads a script, it first translates it into byte code and then it interprets it. If you load a module, you might have noticed a .pyc file. That is the byte code for that module. But you only see these files for modules, ’cause Python doesn’t write the byte code to file for your scripts. For modern virtual machines, like JVM, the byte code is not just interpreted. The machine will analyse how the program executes and compile important functions — the hot spots — into machine code for faster execution. This is what is called just in time compilation (sometimes just too late, but that is a bit cruel). This, by the way, is also the technology running under the hood of Google’s chrome browser. There, the virtual machine runs Javascript, but the idea is essentially the same as for the JVM. Lars Bak, the lead engineer behind that virtual machine, explains it here: To stray a bit from the main point I can mention that I had a class some years ago taught by Lars here in Aarhus. Many of the people now working with Lars had the same class, so that was a successful class. He is going to give it again this year, so don’t you just wish you were studying computer science at our university right now? ;-) Anyway, back to Python. Here you can also get JIT compiling using the module psyco. All you have to is add these two lines to your program: import psyco psyco.full() You can get finer control over the JIT compilation by doing a little more, but this is the simplest way to get it going, and usually it pays off nicely. It is a bit memory hungry, so it isn’t always the thing to do, but more often than not, it is. You get the most out of it when your code is actually doing something serious. Lots of loops and branches and such. Typical algorithmic code. If you are mainly calling built in functions, you do not gain as much. Those are already compiled, typically, so they are pretty optimised as it is. For my scripts, I am mainly scanning through the alignment and updating statistics in a bunch of tables, and I didn’t gain more than maybe a factor of two from psyco. Not really enough for the speed I need. Profiling PythonBefore you go ahead and paint “go faster stripes” on your code, you should always profile it. You will be surprised at where the majority of time is spent. In Python you have three modules for profiling: profile, cProfile and hotshot. The first two have the same functionality, but cProfile is written in C and is faster. I typically use cProfile. The hotshot module is not maintained (according to the Python documentation) and the documentation certainly isn’t, so I have never quite figured out how to use it. I don’t have to either, though, ’cause cProfile does all I need it to. With cProfile you can load the module in and have fine control over what you profile, but I find it the easiest just to profile the entire script. You can do this by loading the module directly when you call your script, like this: python -m cProfile your-script.py After your program has finished, cProfile will write a summary of where the time was spent in the run. If you want to analyse the result in more detail, you can write the profiler’s analysis to a file python -m cProfile -o profile.report your-script.py that you can then read into Python using the module pstats >>> import pstats >>> p = pstats.Stats('profile.report') and you can then sort the report using various criteria. For example, to get the top 10 runtime sinners, you can do: >>> p.sort_stats('time').print_stats(10) Tue Sep 23 15:09:35 2008 profile.report 7932274 function calls in 23.290 CPU seconds Ordered by: internal time List reduced from 119 to 10 due to restriction <10> ncalls tottime percall cumtime percall filename:lineno(function) 1 10.303 10.303 23.276 23.276 extract-statistics.py:2(<module>) 1314684 7.036 0.000 11.282 0.000 extract-statistics.py:47(is_singleton) 5258736 4.246 0.000 4.246 0.000 extract-statistics.py:50(<genexpr>) 377622 1.187 0.000 1.215 0.000 /usr/lib/python2.5/site-packages/bx_python-0.5.0-py2.5-linux-i686.egg/bx/align/core.py:125(column_iter) 862137 0.391 0.000 0.391 0.000 extract-statistics.py:40(is_informative) 9991 0.028 0.000 0.028 0.000 {range} 39960 0.022 0.000 0.022 0.000 /usr/lib/python2.5/site-packages/bx_python-0.5.0-py2.5-linux-i686.egg/bx/align/core.py:193(__init__) 1 0.014 0.014 23.290 23.290 {execfile} 10989 0.014 0.000 0.014 0.000 extract-statistics.py:53(windows) 4995 0.010 0.000 0.013 0.000 extract-statistics.py:28(<genexpr>) Here the topmost line is the entire script, and that is using all the time. Not really a surprise. The next three or four lines tells me where I should put my optimisation efforts. (If you are now wondering why I bother optimising something that takes 23 CPU seconds then congratulations for paying attention to the report and being sceptical about when to bother optimising. I am only running my scripts on a very tiny fraction of my data for the profiling — anything else would take much too long — so that is why. For the real data, optimising really is needed). | ||
| Small World Competition Open for Voting [Bitesize Bio] Posted: 23 Sep 2008 05:30 AM CDT
It looks as though every imaging technique, from photon- or electron-based to computer generated images, has at least one representative entry. I figure that most people will vote for something related to their field (or major). Or am I wrong - is there a particular imaging technique that people find more aesthetic, regardless of background in science? Image: Dr. Eric Hwang at The Scripps Research Institute, La Jolla, CA, USA - Retinoic acid-induced P19 neuronal aggregate (40x) | ||
| Protein evolution switch-off fluorescent proteins (and on) [Reportergene] Posted: 23 Sep 2008 02:39 AM CDT After seven rounds of directed molecular evolution from the RSFP Dronpa, Martin Andresen and colleagues from the Max Plank, report in the September issue of Nature Biotechnology, the generation of two new bright RSFP with unique absorption and switching characteristics. These new proteins, called Padron and bsDrompa will allow for the implementation of monochromatic, multilabeled imaging and dual color far-field fluorescence nanoscopy. Let me give a personal perspective: I bet in a near future someone will exploit the on/off properties of those reporters to get a tomographical solution of fluorescence (and bioluminescence) imaging in living mice. Martin Andresen, Andre C Stiel, Jonas Fölling, Dirk Wenzel, Andreas Schönle, Alexander Egner, Christian Eggeling, Stefan W Hell, Stefan Jakobs (2008). Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy Nature Biotechnology, 26 (9), 1035-1040 DOI: 10.1038/nbt.1493 | ||
| New forum: Next Gen Sequencing Service Providers [SEQanswers.com] Posted: 22 Sep 2008 10:37 PM CDT Per the suggestions of a few people, I've created the to discuss all aspects of third-party next gen sequencing service companies. With the explosion of service providers I've seen in the past year or so, I think this could be a very valuable resource to make sure we're spending our research dollars wisely. So as always...feel free to post questions, reviews, or anything regarding this topic. Read more and join the community... | ||
| Posted: 22 Sep 2008 10:15 PM CDT Emerson looked forward to the day when America would be self-reliant and not second rate in its scholarship. In science, the U.S. has fulfilled Emerson's ambition, but at what cost to religion? Physicist Steven Weinberg muses on religion's fate in the West as science has come to dominate our culture:
| ||
| Quack hunting season is open - update [Bayblab] Posted: 22 Sep 2008 01:32 PM CDT We've talked about how the US authorities have finally started cracking down on bogus cancer cures, including abroad. The FTC has also built a new website to try and warn potential customers about misleading claims and potential hazards of alternative therapies by giving helpful advice: "# Natural doesn't always mean effective. Scammers take advantage of the feelings that can accompany a diagnosis of cancer. They promote unproven – and potentially dangerous – remedies like black salve, essiac tea, or laetrile with claims that the products are both "natural" and effective. But "natural" doesn't mean either safe or effective when it comes to using these treatments for cancer. In fact, a product labeled "natural," can be ineffective and even downright harmful. # Bogus marketers often use trickery and vague language to take advantage of people. Testimonials on websites with ads for products that claim to cure or treat cancer can seem honest and heart-felt, but they can be completely fake: in fact, they may not disclose that actors or models have been paid to endorse the product. Even when testimonials come from people who have taken the product, personal stories aren't reliable as evidence of effectiveness." You can also report bogus cures straight to the FDA, and I know just the one who has been asking for it... Well the list has finally been made public and it includes such gems: "Alexander Heckman d/b/a Omega Supply – Among the products this company marketed are laetrile, which can cause cyanide poisoning when taken orally at high doses; hydrazine sulphate, which is classified by the U.S. Department of Health and Human Services as a potential carcinogen; and cloracesium, which contains celsium chloride. According to the complaint, in addition to making deceptive and false claims that these products are safe and that they effectively prevent, treat, and cure cancer, the respondents also made false claims that the products are scientifically proven to work. Native Essence Herb Company – The products marketed by this company include herbal concoctions (Rene Caisse essiac tea blend and cat's claw), the herb chaparral, and maitake mushrooms extracts. In 1992, the FDA classified chaparral as unsafe because of its "association with acute toxic hepatitis." According to the complaint, the respondents made deceptive and false claims that these products are effective for treating and curing a variety of cancers, eliminating or shrinking tumors, and for preventing breast cancer." Now come and tell me this industry doesn't need regulation... |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |
 There's nothing that hurts soccer practice more than a soccer ball or shoes that went through a pile of dog poo.
There's nothing that hurts soccer practice more than a soccer ball or shoes that went through a pile of dog poo. 






No comments:
Post a Comment