The DNA Network |
| Signal identification [business|bytes|genes|molecules] Posted: 27 Sep 2008 03:28 PM CDT Another great post by Jeff Jonas. The first line says it all
I believe this can be extrapolated to mean that without sufficient and appropriate analytic systems data collection is somewhat meaningless. We have talked a lot about computing everywhere, about research streaming, etc. We haven’t talked enough about filter failure. We also haven’t spent enough time talking about how we can be more effective with the data available to us. The past decade has probably been the triumph of data collection technologies. It’s time for us to harness all that data and get some more meaning from it. Genome sequencing is the obvious example of data generation that leads to new information, and needs to be sliced and diced up as far as we can push it. Michael’s example of research streaming, and my post on XMPP as the foundation for a lab facility, are more along the lines of what Jeff’s talking about and will only lead to a further data explosion, esp in real time data. Having the information is one thing, dealing with it is another. In a streaming data, or high volume data scenario, we need to pay attention to items #1 and #2 in Jeff’s list, Favoring the false negative, and most importantly, doing a better job of calculating relevance. His use case is very different from what I am talking about here. We are not calculating risk, but risk calculation is not too different from developing a genomic disease signature, or trying to figure out what patterns we might want to capture from our experiments. Related articles by Zemanta |
| Are your sperm short-lived and tiny? [Discovering Biology in a Digital World] Posted: 27 Sep 2008 03:16 PM CDT Then you need to head over to The Oyster's Garter and read Miriam Goldstein's incredibly funny post about the problems of male sea squirts. Apparently, if you're a sea squirt, the size of your sperm is related to your environment and mostly the number of other sea squirts in your environment. Usually, I'm not attuned to the issues of broadcasting spawn or the lives of those who live under a dock, but Ms. Goldstein channels Dr. Tatiana Sometimes extremely handsome pleats just aren't enough. Read the comments on this post... |
| Humorous sciency signs #1: Please do not molest the spawning salmon [The Tree of Life] Posted: 27 Sep 2008 03:05 PM CDT |
| Posted: 27 Sep 2008 01:03 PM CDT One of the holy grails of modern medicine is the development of a vaccine against HIV, the virus that causes AIDs. An obstacle to attaining this goal has been the difficulty in stimulating the immune system to make it produce the right kinds of antibodies. A recent finding in Science describes a gene that controls production of these antibodies and may provide insights to the development of an effective vaccine. (1). |
| Abstracts From the ASHG 2008 Meeting [The Genetic Genealogist] Posted: 27 Sep 2008 08:48 AM CDT Interestingly, the first five abstracts all include researchers from the Sorenson Molecular Genealogy Foundation, showing how much the Foundation is providing to the genetic genealogy community. Also very interesting is the final abstract which argues that genetic genealogy, in combination with large-scale genomic analyses, will result in reduced privacy.
Allocation of YSTR Microvariant Alleles to Y-Chromosome Binary Haplogroups. A. L. Pollock, K. Ritchie, P. A. Underhill, A. A. Lin, S. R. Woodward, U. A. Perego, N. M. Myres "To identify YSTR microvariant alleles potentially useful for elucidating further phylogenetic substructure within binary haplogroups, we have assessed the haplogroup affiliation of microvariant alleles found at informative frequencies in public YSTR databases for the following YSTR loci: DYS385, DYS392, DYS441, DYS446, DYS447, DYS449 and DYS464. We report haplogroup affiliations for each variant allele and geographic origins of representative samples." Read more here… L1c2a, the (African) Haplogroup With The Longest Mitochondrial Genome! K. Ritchie, U. A. Perego, A. Achilli, N. Angerhofer, N. M. Myres, A. Torroni, S. R. Woodward "During a recent survey of the nearly 58000 mtDNA control region haplotypes currently present in the publicly accessible Sorenson Molecular Genealogy Foundation database, we observed a small number of mtDNAs (n=16) characterized by the presence of unusually long insertions of up to 200 bases. A small subset of these particularly long mtDNA haplotypes shared an identical insertion of 15 bases." Read more here… The mitochondrial DNA landscape of modern Mexico. A. Achilli, U. A. Perego, J. E. Gomez-Palmieri, R. M. Cerda-Flores, K. H. Ritchie, A. Pollock, N. Angerhofer, A. Escobar-Mesa, A. Torroni, N. M. Myres, S. R. Woodward, Sorenson Molecular Genealogy Foundation, SLC, Utah (USA) "Analysis of the mitochondrial DNA (mtDNA) control region sequences, including HVS-I, HVS-II and HVS-III, from more than 2,000 subjects revealed an overwhelming Native American legacy in the modern Mexican population, with ~90% of mtDNAs belonging to the four major pan-American haplogroups A2, B2, C1 and D1. This finding supports a European contribution to the Mexican gene pool primarily by male settlers and confirms the effectiveness of employing the uniparentally-transmitted mtDNA as a tool to reconstruct a country's history." Read more here… The origin of Native Americans from a mitochondrial DNA viewpoint. U. A. Perego, A. Achilli, L. Milani, M. Lari, M. Pala, A. Olivieri, B. Hooshiar Kashani, J. E. Gomez-Palmieri, N. Angerhofer, A. Pollock, K. H. Ritchie, N. M. Myres, S. R. Woodward, D. Caramelli, A. Torroni "Our comprehensive overview of the four pan-American branches of the mtDNA tree suggests a scenario with a human entry and spread into the Americas from Beringia about 20,000 years ago, and preliminary data raise the possibility that the uncommon five Native American haplogroups might have marked additional migratory events from Asia or Beringia. Overall, through a combined analysis of modern and ancient Native American mtDNA, we are making an effort for reconstructing the complex pre-Columbian history at both macro- and micro-geographic levels." Read more here… Mitochondrial DNA footprints in modern Mongolia. S. R. Woodward, A. Achilli, U. A. Perego, J. E. Gomez-Palmieri, D. Tumen, E. Myagmar, D. Bayarlhagva, K. H. Ritchie, A. Pollock, N. Angerhofer, A. Torroni, N. M. Myres, Sorenson Molecular Genealogy Foundation, SLC, UT (USA) "In 2007, through a well-planned collection effort, researchers at the Sorenson Molecular Genealogy Foundation and the National University of Mongolia were able to gather over 3,000 DNA samples, informed consents, and genealogical data throughout the country of Mongolia, including samples from 21 distinct tribal or ethnic populations. All the samples were sequenced for the three hypervariable segments of the mitochondrial DNA (mtDNA) control region to assess the genetic composition of modern Mongolia." Read more here… Early Siberian Maternal Lineages in the Tubalar of Northeastern Altai Inferred from High-Resolution Mitochondrial DNA Analysis. R. Sukernik, I. Mazunin, E. Starikovskaya, N. Volodko, N. Eltsov "We showed that the core of the Tubalar genetic makeup proved to be a mixture of "west" (H8, U4b, U5a1, and X2e) and "east" Eurasian (A and B1) haplogroups derived from macrohaplogroup N, and Siberian derivatives of the macrohaplogroup M identifiable by subhaplogroup-specific mutations. For example, among the 36 Tubalar mtDNA samples that belong to haplogroup D, 10 (28%) harbored diagnostic markers of the subhaplogroup D3a2a shared with the Chukchi and Eskimos. This finding verified at the complete sequence level we attributed to ancient link between early Siberians, who underwent pronounced differentiation in the Altai-Sayan region, and some of the Eskimo tribes." Read more here… Population Structure in Mongolia from a Mitochondrial DNA Perspective. L. Pipes, A. A. Pai, D. Labuda, T. G. Schurr "To clarify the complex population history of Mongolia, we analyzed variation in the mtDNAs of 190 individuals from several Mongolian ethnic groups, including the Uriankhai, Zakhchin, Derbet, Khoton and Khalkha. We screened all samples for phylogenetically informative coding region SNPs and sequenced HVSI to assess control region variation in them. Our data suggest that the mtDNA diversity present in our population is consistent with the general pattern of variation observed in East Asia, with the most frequent haplogroups being C, D and G. Haplogroup variation in Mongolian ethnic groups reveals considerable maternal diversity with a predominance of basal M types. Interestingly, the Mongolians also possessed West Eurasian haplogroups, such as H, J and K, which are not commonly observed in East Asia, even at low frequencies. Read more here… Genetic History of human populations of East African inferred from mtDNA and Y chromosome analyses. J. Hirbo, S. Omar, M. Ibrahim, S. Tishkoff “Our results indicate that East African populations have some of the most ancestral Y chromosome and mtDNA lineages in Africa, suggesting that they may have been an ancient source of dispersion throughout Africa. Additionally, we find evidence for ancient geneflow between East Africa and the Middle East. We also ascertained the effect of the Bantu-expansion and signature of recent migration of Cushitic-speaking groups originating from Ethiopia on peopling of East Africa.” Read more here… Analysis of mtDNA and Y-chromosome haplogroups in Mexican Mestizos and Amerindian groups. I. Silva-Zolezzi, B. Z. Gonzalez-Sobrino, J. K. Estrada-Gil, A. Contreras, J. C. Fernandez, E. Hernandez-Lemus, L. Sebastian, F. Morales, R. Goya, C. Serrano, G. Jimenez-Sanchez "For this we included genotypic data from 163 mt SNPs and 123 Y chromosome SNPs present in the Illumina Human1M chip of 450 individuals, 300 mestizos from six states located in different regions: Northern, Central and Southern; and 150 individuals from different Amerindian groups (Tepehuanes, Zapotecos and Mayas). With this information, we are measuring genetic diversity using Fst and AMOVA analysis. Admixture analysis includes average and individual ancestral contribution estimates using autosomal SNPs. Initial results show that in our Mestizo sample, 88% of the mt haplogroups are Amerindian (A, B, C or D), and the rest includes European and African lineages. We have identified differences in proportions of each haplogroup in both Mestizos and Amerindians." Read more here… Using mtDNA and Y-chromosome for estimating group ancestry: Implications for case-control studies. K. Stefflova, M. Dulik, A. Pai, A. Walker, T. Schurr, T. Rebbeck “We examined the possible role of mtDNA and the non-recombining portion of the Y-chr. (NRY) as ancestry informative markers (AIMs) for admixed groups (self-identified African Americans (AA) or European Americans (EA)) collected as part of a prostate cancer case-control study. We deeply typed both mtDNA (HVS-I, II, 36 coding SNPs) and the NRY (37 SNPs) in a group of 226 AA cases and controls and compared this group to 206 EA cases and controls, and 49 Senegalese…We found a sex biased admixture for AA where 13.2% of mtDNAs and 34.5% of NRYs were of non-African origin. We also found a small amount of admixture in EA (~3% mtDNA, 1.5% NRY).” Read more here… New tool (mtPHYL) proposed for phylogenetic analysis of human complete mitochondrial genomes. N. Eltsov, N. Volodko, E. Starikovskaya, R. Sukernik Y chromosome microsatellite haplotypes in the Hutterite founders. M. Caliskan, I. Pichler, C. Platzer, P. P. Pramstaller, C. Ober "The current population of >12,000 Schmiedeleut Hutterites are descendants of 38 male founders who were born between 1700 and 1830 in Europe. Only 12 of these founders, each with a unique surname, have living male descendants related through male-only lineages. DNA samples were available in our laboratory for 75 male descendants of 11 of the 12 founders, accounting for 673 independent paternal meioses. We genotyped 9 microsatellite loci, which included a mean of 6.8 (range 2-23) males per lineage to evaluate potential relationships between the founders. Fourteen different haplotypes were identified, with an average of 3.5 (range 1-8) pairwise differences between haplotypes. All descendants within each of 9 lineages had identical Y haplotypes. Descendents of two of these lineages, 2 and 10, had the same haplotype despite different surnames, suggesting possible relatedness between the founders of these two lineages." Read more here… Genetic variation in tribes of Eastern and North-Eastern India: inference from distribution of Y-chromosomal polymorphisms. M. Borkar, F. Ahmed, F. Khan, S. Agrawal Inferential Genotyping in Mormon Founders and Utah pedigrees. J. Gitschier |
| The ties that bind [genomeboy.com] Posted: 27 Sep 2008 07:49 AM CDT
Oh really? A bridge you say? |
| What’s on the web? (27 September 2008): Ping pong balls and forecasting the future [ScienceRoll] Posted: 27 Sep 2008 04:42 AM CDT
 |
| HelixGene Foundation: THE Genomic Resource [ScienceRoll] Posted: 27 Sep 2008 04:12 AM CDT Steve Murphy, our gene sherpa and the clinical genetics fellow at Yale University, came up with something great again. The Helix Foundation is up and running now.
They are so right! When you hear about articles mentioning scientists who just found the gene of obesity or intelligence, physicians tend to trust this kind of information. Now hopefully, they will have a fantastic source to rely on. The first step is giving more details about the misinformation regarding Sergey Brin’s risk for Parkinson’s disease. I believe that one was a marketing trick, nothing more. Other bloggers seem to agree with me on this:
 |
| Python lectures [Mailund on the Internet] Posted: 27 Sep 2008 04:00 AM CDT |
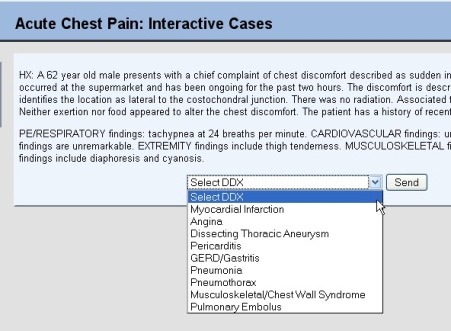
| ACDET: Diagnosis Training [ScienceRoll] Posted: 27 Sep 2008 03:50 AM CDT I just came across a useful training tool at ACDET:
You get a patient’s history then decide which diagnosis to choose. We need tools like this in our community, the Medical Education Evolution.  |
| Meet a cancer survivor in Second Life! [ScienceRoll] Posted: 27 Sep 2008 02:34 AM CDT First, there will be a medical exercise/intern meeting today at the Ann Myers Medical Center in Second Life at 10:00 SL time. (Teleport link) Topic: marburg. And you can also find a poster at the AMMC about an event organized by the American Cancer Society. Meet a cancer survivor (Child hood Leukemia) on the 28th of September at noon SL time. Details on the poster.  |
| Bella star urges pro-life vote [Mary Meets Dolly] Posted: 26 Sep 2008 08:58 PM CDT I do not know exactly what Eduardo Verástegui is saying, but I don't care. Not only is he a gorgeous man and a talented actor, but he is pro-life, and according to the translation he is urging Hispanic voters to cast their ballots for life. I think I am in love... |
| Attention, science and money [business|bytes|genes|molecules] Posted: 25 Sep 2008 10:39 PM CDT Interesting observation by Kevin Kelly. He says
To some extent, that’s somewhat obvious. Peter Drucker, whom I admire a lot, said the following
Attention can be driven by many mechanisms, marketing being the most effective one. The key is gaining sufficient mindshare, which is often accompanies by a flow of capital. In science, the money follows topics of research that have mindshare. Similarly people fund companies in areas that generate mindshare for whatever reason. The question I often ask myself, both from my time as a marketer and as someone interested in science communication, is how can we bring more mindshare to some of our efforts and science in general. What does money flow mean? Is it just research funding? Is it investment in such concepts as “bursty work”? Take something else Kelly writes
I have written about the lack of marketing in science (stealing shamelessly from Larry Page). It’s critical that we do a better job of highlighting the power of our activities and learn some marketing tricks along the way. No I am not talking about the in your face stuff that gives marketing a bad name, but about the kinds of activities that maintain that attention, and get people to notice. The good news, many of us already do that, perhaps without even realizing it. It’s still niche awareness, but I have a feeling that we are close to actually crossing the hump and bringing some of our activities into the mainstream. KK link via Michael Nielsen |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |
![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_b.png?x-id=877db394-c289-4c8f-a5e9-52ee14bd353a)




![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_b.png?x-id=776b9a7b-c14f-45c1-8800-0b470237ffe7)
No comments:
Post a Comment