The DNA Network |
| Science and polittics: Canadian elections [Bayblab] Posted: 18 Sep 2008 06:23 PM CDT While the world watches the run-up to the US election in a background of near economic collapse few noticed that Canadians will also go to the polls this fall. Canadian's themselves seem to be paying attention to the south more than their own backyard (66% favor Obama, 14% McCain). While there was some coverage of science issues in the US election we have yet to hear much about it in Canada. A quick look at the campaign sites of the 5 major parties shows that all focus to some degree on global warming but don't delve much deeper. Liberals are pushing their "green shift" but don't pay much attention to other science issues. Conservatives don't seem to have a definite plan but like to boast about their environmental record. The NDP mentions both environmental protection and the increasingly popular cap-and-trade system. The green party is not-surprisingly the most thorough. The Bloc Quebecois have a nice video. So over the next weeks we'll try to see how the different parties stack up on science issues. Yes the environment is a particularly pressing one, but there are many others: science funding, reinstating a scientific advisor etc... leave it to Nature, a British journal, to have the best editorial and news on the issue so far... follow the link for more reading: Indeed, many Canadian scientists are seeing, and complaining about, an undue emphasis on commercially focused research over long-term basic research. Such complaints are heard in many other countries too. But in Canada the problem is compounded by the fact that the current government has channelled new science funds into four restrictive priority areas — natural resources, environment, health and information technology — and that scientists are often required to scrounge matching funds from elsewhere to top up their grants. Furthermore, the government this month defined sub-priority areas that mix in obvious commercial influences: alongside 'Arctic monitoring', for example, sits 'energy production from the oil sands'. |
| Search-and-Destroy mission for Down Syndrome fetuses effects normal fetuses too [Mary Meets Dolly] Posted: 18 Sep 2008 05:48 PM CDT UK doctors are estimating that two normal fetuses are miscarried for every three Down syndrome fetuses that are detected. Here is a perfect example of genetic testing gone horribly wrong. Genetic testing can be a positive thing, except when it is used for a "search-and-destroy" mission. From the Telegraph:
"Prevented from being born" Wow. That certainly is a nice way of saying "ripped out of mother's womb and killed for having an extra chromosome." Hat Tip: Wesley J. Smith |
| Human cloning: the child of IVF [Mary Meets Dolly] Posted: 18 Sep 2008 05:27 PM CDT The Catholic Church is opposed to in vitro fertilization, better known as IVF. This opposition is not a "punishment" for infertile couples but an affirmation of the sanctity of human life. The Church knew in the 1970's, as it does today, that once procreation was taken out of the womb and children were created in laboratories, that human life would become a commodity. Human life would no longer be a gift from God but a product of technical intervention. Human life that was "begotten not made", would no longer be "begotten" but would become "man-made". And sure enough, human embryos created by IVF are now a valuable commodity and considered by many to be simply harvestable biological material. It doesn't stop there. Once creating human lives outside the womb is acceptable, then human cloning is the next logical step. IVF has begotten human cloning. Don't believe me? This story proves the link between IVF and human cloning. And this clinic is by no means unique. From Domain-B.com:
Now it is therapeutic cloning, or cloning-to-produce stem cells. But, when reproductive cloning, or cloning-to-produce children, becomes a reality, it will be the IVF clinic that offers it as part of their "reproductive choices" menu. |
| New theme [Mailund on the Internet] Posted: 18 Sep 2008 05:22 PM CDT I want a simpler theme on my blog, so while I was sitting here waiting for some computations to finish, I have been playing around with themes. This is what I came up with. It is much simpler to look at than the old theme. That’s the good part. To get to this, I’ve had to remove some of the widgets from the side bars that I usually like. Such as the DNA Network image showing the resent headlines. I liked that one, but it had to go to get a cleaner theme. Instead I’ve added a blogroll so I can check up on e.g. the DNA Network through that. Now my computations are done, so I’ll have a quick look at it and start up the next batch. Then it’s off to bed. I’m getting up in 5 hours… |
| Paper: "Rapid whole-genome mutational profiling using next-gen technolgies" [SEQanswers.com] Posted: 18 Sep 2008 05:07 PM CDT |
| Trans-splicing and Chromosomal Translocations [Bayblab] Posted: 18 Sep 2008 04:07 PM CDT  I'm up for the research group journal club tomorrow. Since a couple of the bayblab readers will not be able to make it, I thought I'd pass on what I'll talk about. A recent publication in Science, demonstrates that an mRNA in a normal endometrial cells encodes the exact same fusion protein that normally occurs as the result of a translocation found in 50% of endometrial cancers. The transcript, as detected by RT-PCR and RNase protection assay is found in abundances that are affected by hormone treatment and hypoxia, suggesting that it has a biological role. In cancer this fusion protein results in resistance to apoptosis and increased proliferation. However, in theses normal cells, there is no DNA encoding for this hybrid mRNA and the authors suggest it is a result of trans-splicing. Trans-splicing of this sort has not been observed in vertabrate cells and even in other organisms it is quite different. They go to great pains, of course, to demonstrate that it is not an RT-PCR artifact and that there is not a subpopulation of the normal cells that contain the translocation. The killer is that they do in vitro trans-splicing assays using Rhesus and Human nuclear extracts and indeed find this chimeric mRNA. I'm sure they are looking for other fusion mRNAs using this assay in a high throughput method. This study raises some intersting question about these oncogenic fusion proteins that are very common in cancer. Particular fusion proteins are repeatedly found in particular cancers. Do they have a function in normal cells? Is it through the mechanism that produces these mRNAs or the mRNAs themselves that cause these cancer associated translocations?? At least check out the figure. Translocation is indicated in the centre of the figure. Two mRNAs combine to create a novel one that has no DNA equivalent. |
| Has Science Really Been Under Lock and Key? [adaptivecomplexity's column] Posted: 18 Sep 2008 02:00 PM CDT A current news piece announces that the end of scientific secrecy. Scientists can "forgo the long wait to publish in a print journal and instead to blog about early findings and even post their data and lab notes online. The result: Science is moving way faster and more people are part of the dialogue." Really? |
| The origin of evolution [Bayblab] Posted: 18 Sep 2008 01:51 PM CDT I found this rather neat paper in PNAS on how natural selection and evolution actually predate the origin of life. If you assume that to be alive you must require the conditions of being able to replicate and evolve, than you have to wonder which came first. According tho their model, not only did abiotic information systems undergo natural selection but evolution favored the emergence of replication. It's a short read if you understand mathspeak (which I don't), but here's the gist of it from the concluding paragraph.... Traditionally, one thinks of natural selection as choosing between different replicators. Natural selection arises if one type reproduces faster than another type, thereby changing the relative abundances of these two types in the population. Natural selection can lead to competitive exclusion or coexistence. In the present theory, however, we encounter natural selection before replication. Different information carriers compete for resources and thereby gain different abundances in the population. Natural selection occurs within prelife and between life and prelife. In our theory, natural selection is not a consequence of replication, but instead natural selection leads to replication. There is ''selection for replication'' if replicating sequences have a higher abundance than nonreplicating sequences of similar length. We observe that prelife selection is blunt: Typically small differences in growth rates result in small differences in abundance. Replication sharpens selection: Small differences in replication rates can lead to large differences in abundance |
| What do you mean by "race"? A call for standards and empirical ethics research. [PredictER Blog] Posted: 18 Sep 2008 01:27 PM CDT You've probably read an article or two that reports the results of a race-based investigation. Perhaps it was a study of health disparities, a survey of patient attitudes, an examination of a race-based intervention or new medication to be marketed to a specific race-based demographic. If you wanted to do a systematic review of these papers, you might be vexed by the difficulty of finding a common, valid definition of "race". As a socially-constructed categorie, defining the limits of race and ethnicity is a slippery business and one that has a problematic past in the history of science and medicine. With this in mind, Vural Ozdemir, Janice E. Graham and Beatrice Godard make a call for clarity in "Race as a variable in pharmacogenomics science: from empirical ethics to publication standards" (Pharmacogenet Genomics. 2008 Oct;18(10):837-41. - PubMed | CiteULike). The authors argue for the use of empirical ethics research to inform the development of new publication standards to "minimize the drift from descriptive to attributive use of race in publications". In this context, empirical ethics, or "applied social science methodologies … to better understand, for example, the 'lived' experiences of user groups", would identify blind-spots in predictive health research and would help researchers, regulators, policy-makers, and editors "differentiate between an imprecise (yet measurable) predictive biomarker, from a construct such as race". Given the uproar around BiDiL and other race-based pharmacogenomic ventures, the authors have made a timely, if not over-due, call for publishers and ethics researchers to collaborate in developing standards for the use of the controversial category in published research. - J.O. |
| Road Trip: 454 Users Conference [FinchTalk] Posted: 18 Sep 2008 01:25 PM CDT |
| do.call the lifesaver [Mailund on the Internet] Posted: 18 Sep 2008 11:21 AM CDT It is not uncommon for me to summarize some data in a set of files — say one per chromosome in a genome analysis — and then want to read the entire data into R for analysis. This is a matter of calling read.table() on each file and then combining the results with rbind(), and not really a major problem, but I’ve never been happy with the way I did it. I would iterate through the file names and append the tables together one at a time, something that can be very slow when there is a lot of files and a lot of data. Today I stumbled upon the function do.call() in the documentation. It is essentially like apply in Python, and just what I need to solve this problem. Now, to read in tables from a bunch of files I can use simply: data <- do.call("rbind",lapply(fnames,read.table,header=TRUE)) Neat, eh? |
| Rapid NJ [Mailund on the Internet] Posted: 18 Sep 2008 10:24 AM CDT Our new neighbour-joining paper (I wrote about it here, but it was called “Accelerated Neighbour-Jouining” then) just came online:
I have put the source code plus binaries for Linux (Debian and RedHat), Windows and Mac OS X (i386) on this web page. |
| Release of CLC Genomics Workbench 2.0 [Next Generation Sequencing] Posted: 18 Sep 2008 09:29 AM CDT Here at CLC bio the developer team is really proud to have released the second major version of our Genomics Workbench. A full list of the new functionalities is provided here. Most importantly the release includes: Support for reference assembly against the human genome (i.e. reference sequences of any size) New and much faster algorithm for assembling short reads [...] |
| Goldstein redux [genomeboy.com] Posted: 18 Sep 2008 07:55 AM CDT I can tell when a science story has created mainstream buzz: I read about it here. The blogosphere has reacted–with nuance and thoughtfulness, IMHO– to David Goldstein’s contention that the Common Disease Common Variant Emperor is pretty much buck naked. In my own inbox over the last two days, I’ve seen loud and emphatic grumblings from NIH-funded researchers who claim that Goldstein is prematurely eulogizing CDCV and that if he hasn’t found variants related to cognition, then it must be the result of poor phenotyping on his part. Fiddlesticks. I am a subject in Goldstein’s cognition study and I can tell you that I have been tested out the wazoo. I have spent hours in front of a screen trying to remember patterns of dots, letters and numbers. I have sat with an examiner for an hour and tried to recall the details of stories that were read to me. I have lain inside an MRI tube while staring at photographs and answered questions about them. I have filled out lengthy questionnaires. So yeah, it could be inadequate phenotyping, but if it is then I suspect we will never have adequate phenotyping until we start drilling holes in people’s skulls. (Also, if the powers that be are so concerned about this, then why isn’t the 1000 Genomes Project using some of its $50 million to collect trait data? Where is the 1000 Phenomes Project?) As for the common-disease-common-variant hypothesis, maybe I’m missing something, but please tell me: What do we do with so many weak susceptibility loci for Crohn’s and type 2 diabetes that fail to explain so much of the genetic variance? Schizophrenia has a heritability of 0.8 and we still can’t find a major gene after 25 years of looking. Uncle Sam and his fundees have a lot invested in CDCV and the genome-wide association studies that are supposed to find the culpable variants for human diseases. I don’t blame them for wanting to pursue it as far as they can. But when will it be time to move on and start to sequence? I get that it’s still expensive and unwieldy. And that it too may not succeed. But why use a magnifying glass when you have an electron microscope? UPDATE: I just heard an excellent talk by Muin Khoury (some of which irritated me, but we can talk about that later). One of his many salient points was that CDCV has not born fruit because we have failed to take into account gene-gene and gene-environment interactions. This strikes me as correct and more importantly, as the source of an immense if not intractable informatics and computational problem. Of course, someone more cynical than I might say that it will also mean many more years of funding… |
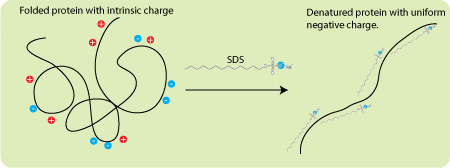
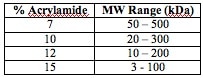
| How SDS-PAGE works [Bitesize Bio] Posted: 18 Sep 2008 04:18 AM CDT SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) is commonly used in the lab for the separation of proteins based on their molecular weight. It’s one of those techniques that is commonly used but not frequently fully understood. So let’s try and fix that. SDS-PAGE separates proteins according to their molecular weight, based on their differential rates of migration through a sieving matrix (a gel) under the influence of an applied electrical field. Making the rate of protein migration proportional to molecular weight. The movement of any charged species through an electric field is determined by it’s net charge, it’s molecular radius and the magnitude of the applied field. But the problem with natively folded proteins is that neither their net charge nor their molecular radius is molecular weight dependent. Instead, their net charge is determined by amino acid composition i.e. the sum of the positive and negative amino acids in the protein and molecular radius by the protein’s tertiary structure. So in their native state, different proteins with the same molecular weight would migrate at different speeds in an electrical field depending on their charge and 3D shape. To separate proteins in an electrical field based on their molecular weight, we need to destroy the tertiary structure by reducing the protein to a linear molecule, and somehow mask the intrinsic net charge of the protein. That’s where SDS comes in. The Role of SDS (et al) SDS is a detergent that is present in the SDS-PAGE sample buffer where, along with a bit of boiling, and a reducing agent (normally DTT or B-ME to break down protein-protein disulphide bonds), it disrupts the tertiary structure of proteins. This brings the folded proteins down to linear molecules. SDS also coats the protein with a uniform negative charge, which masks the intrinsic charges on the R-groups. SDS binds fairly uniformly to the linear proteins (around 1.4g SDS/ 1g protein), meaning that the charge of the protein is now approximately proportional to it’s molecular weight. SDS is also present in the gel to make sure that once the proteins are linearised and their charges masked, they stay that way throughout the run. The dominant factor in determining an SDS-coated protein is it’s molecular radius. SDS-coated proteins have been shown to be linear molecules, 18 Angstroms wide and with length proportional to their molecular weight, so the molecular radius (and hence their mobility in the gel) is determined by the molecular weight of the protein. Since the SDS-coated proteins have the same charge to mass ratio there will be no differential migration based on charge. As the name suggests, the gel matrix used for SDS-PAGE is polyacrylamide, which is a good choice because is chemically inert and, crucially, can easily be made up at a variety concentrations to produce different pore sizes giving a variety of separating conditions that can be changed depending on your needs. You may remember that I previously wrote an article about the mechanism of acrylamide polymerisation previously - click here to read it. To conduct the current through from the anode to the cathode through the gel, a buffer is obviously needed. Mostly we use the discontinuous Laemmli buffer system. “Discontinuous” simply means that the buffer in the gel and the tank are different. Typically, the system is set up with a stacking gel at pH 6.8, buffered by Tris-HCl, a running gel buffered to pH 8.8 by Tris-HCl and an electrode buffer at pH 8.3. The stacking gel has a low concentration of acrylamide and the running gel a higher concentration capable of retarding the movement of the proteins. So what’s with all of those different pH’s? Well, glycine can exist in three different charge states, positive, neutral or negative depending on the pH. This is shown in the diagram below, which is taken from an excellent tutorial that explains the whole thing. Control of the charge state of the glycine by the different buffers is the key to the whole stacking gel thing. So here’s how the stacking gel works. When the power is turned on, the negatively-charged glycine ions in the pH 8.3 electrode buffer are forced to enter the stacking gel, where the pH is 6.8. In this environment glycine switches predominantly to the zwitterionic (neutrally charged) state. This loss of charge causes them to move very slowly in the electric field. The Cl- ions (from Tris-HCl) on the other hand, move much more quickly in the electric field and they form an ion front that migrates ahead of the glycine. The separation of Cl- from the Tris counter-ion (which is now moving towards the cathode) creates a narrow zone with a steep voltage gradient that pulls the glycine along behind it, resulting in two narrowly separated fronts of migrating ions; the highly mobile Cl- front, followed by the slower, mostly neutral glycine front. All of the proteins in the gel sample have an electrophoretic mobility that is intermediate between the extreme of the mobility of the glycine and Cl- so when the two fronts sweep through the sample well the proteins are concentrated into the narrow zone between the Cl- and glycine fronts. And they’re off! This procession carries on until it hits the running gel, where the pH switches to 8.8. At this pH the glycine molecules are mostly negatively charged and can migrate much faster than the proteins. So the glycine front accelerates past the proteins, leaving them in the dust. The result is that the proteins are dumped in a very narrow band at the interface of the stacking and running gels and since the running gel has an increased acrylamide concentration, which slows the the movement of the proteins according to their size, the separation begins. What was all of that about? If you are still wondering why the stacking gel is needed, think of what would happen if you didn’t use one. Gel wells are around 1cm deep and you generally need to substantially fill them to get enough protein onto the gel. So in the absence of a stacking gel, your sample would sit on top of the running gel, as a band of up to 1cm deep. Rather than being lined up together and hitting the running gel together, this would mean that the proteins in your sample would all enter the running gel at different times, resulting in very smeared bands. So the stacking gel ensures that all of the proteins arrive at the running gel at the same time so proteins of the same molecular weight will migrate as tight bands. Separation Once the proteins are in the running gel, they are separated because higher molecular weight proteins move more slowly through the porous acrylamide gel than lower molecular weight proteins. The size of the pores in the gel can be altered depending on the size of the proteins you want to separate by changing the acrylamide concentration. Typical values are shown below. For a broader separation range, or for proteins that are hard to separate, a gradient gel, which has layers of increasing acrylamide concentration, can be used. I think that’s about it for Laemmli SDS-PAGE. If you have any questions, corrections or anything further to add, please do get involved in the comments section! |
| Street View on the cellphone [Mailund on the Internet] Posted: 18 Sep 2008 12:19 AM CDT Google’s at it again. A new and improved Google Maps for the mobile phone. I can see how this can be useful when travelling.
Now, my phone doesn’t have GPS or any other way of tracking its location, but I wonder if the Maps keep track of where you are. Does anyone know? |
| Warfarin Sensitivity, Iverson Genetics, and Personalized Medicine on NPR [DNA Direct Talk] Posted: 17 Sep 2008 04:04 PM CDT Last week, NPR's "Morning Edition" did a story on the way companies and entrepreneurs are beginning to develop and market products tailored to the genetic makeup of individual patients. Notably, the story featured Iverson Genetic Diagnostics and their test for warfarin sensitivity. Warfarin (brand-name Coumadin®) is a commonly used blood-thinner, but doctors often have a hard [...] |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |


No comments:
Post a Comment