The DNA Network |
| How Bacteria Recognize Kin [adaptivecomplexity's column] Posted: 12 Jul 2008 05:50 PM CDT A report in this week's issue of Science describes a set of genes that enable members of the bacterial species Proteus mirabilis to tell the difference between kin and strangers. The bacteria engage in a social behavior (common to teenagers and bacteria) called swarming: they like to get together in groups. If you spread these bacteria on a petri dish, they are able to move together to form a colony. The catch is this: like humans, bacteria of the same species can be divided up into smaller populations of more closely related individuals. |
| Posted: 12 Jul 2008 04:03 PM CDT You may have heard the news that Louisiana's governor recently signed an "Academic Freedom" bill, the first such bill to pass in a recent string of efforts to allow public school teachers to push non-scientific alternatives to evolution. (I previously wrote about Missouri's failed version.) All of these bills claim to promote academic freedom for public school teachers to teach the Intelligent Design movement's so-called evidence against evolution. But the concept of academic freedom in a high school curriculum makes no sense. In the New Scientist story linked to above, Josh Rosenau of the National Center for Science Education points out that "if you look at the American Association of University Professors' definition of academic freedom, it refers to the ability to do research and publish." The whole point of academic freedom is, like tenure, to protect independent scholars and scientists from having their work suppressed, manipulated, or managed by administrators or other people outside the research community who might want to pressure scholars to alter their conclusions or not research unfavorable topics. |
| Climate Change and the Future of California's Wildflowers [Tomorrow's Table] Posted: 12 Jul 2008 02:08 PM CDT California is burning. The smoke obscures my view of Lake Tahoe from our cabin on the eastern side of the Sierra Nevada Mountains. I can also no longer see the distinctive snow cross on the flank of the 10,000-foot peak of Mt Tallac. The children cough and I rub my eyes. The smokiness invades our cabin, our clothes, our hair. Ash drops out of the sky. It is hot. Again? Just last summer, the Angora fire burned within 4 miles of our cabin. This June, a massive lightning storm sparked 800 wildfires across the state- one of the largest is burning in the foothills about 100 miles from here. Perhaps I shouldn't be so surprised. After all, heat and fire are predicted consequences of global climate change. Officials said the unprecedented fire season, fuelled by drought and 100 degrees Fahrenheit temperatures, had seen the most fires burning at any one time in recorded California history. I see it, I smell it. I breathe it. I get it. Discouraged, I walk barefoot outside to check on one of my favorite wildflowers- the Explorers gentian (Gentiana calycosa). It grows in a small meadow at the base of a massive incense cedar (Calocedrus decurrens). I see that the buds are just forming on the low leafy plants. Later this summer, the Gentian will regale us with exquisite showy masses of bright blue, yellow-spotted corolla.  I continue to the back of the house to a wooded thicket, where one-sided wintergreen (Pyrola picta), a small, evergreen perennial grows. Small and unobtrusive, with its flower buds perched awkwardly on one side of its single flowering stock, this tiny wintergreen seems as if it is to shy to join the party. But up close, I can see that this is no wallflower. Its buds are bell-shaped with beautiful pale green flowers.  Down the hill, under the pines, I see a patch of bright red. This is the solitary snow plant (Sarcodes sanguinea). Always the first flash of color after the snow melt, this stout fleshy plant is the joy of the children. It lacks chlorophyll so cannot make its own food through photosynthesis. Apparently it supplements its nutrient intake by at least partially parasitizing the roots of pine trees by means of a shared mycorrhizal fungus.  My favorite plants, those that have brought joy to my family each summer for over 50 years, appear to be fine. I am reassured. But what will the climate change do to these species? Now we have an answer and it is not good. In a recent article in PLoS One, Loarie et al., assess the potential impacts of climate change on the native flora of California. They examine 8 different potential scenarios for the future of the California flora in the face of climate change. They project that up to 66% of the California native species will experience >80% reductions in range size within a century. They find that the foothills of the northern Sierra Nevada are extremely vulnerable to species loss and that species in some mountainous areas will shrink in range. These areas include the Gentian, the snow plant and the wintergreen. Photos courtesy of Matt Below. Loarie, S.R., Carter, B.E., Hayhoe, K., McMahon, S., Moe, R., Knight, C.A., Ackerly, D.D., McClain, C.R. (2008). Climate Change and the Future of California's Endemic Flora. PLoS ONE, 3(6), e2502. DOI: 10.1371/journal.pone.0002502 |
| Science Webinar: CNVs vs SNPs [ScienceRoll] Posted: 12 Jul 2008 01:37 PM CDT Science Magazine will organize a webinar about copy number variations and single nucleotide polymorphisms.
Speakers:
The moderator will be Sean Sanders, Ph.D., Commercial Editor, Science/AAAS.  |
| Industry Watching: Pharma looks at new ways to innovate [business|bytes|genes|molecules] Posted: 12 Jul 2008 01:21 PM CDT
The scientific weight behind Enlight makes an impressive list, as does the list of pharma partners, and the areas of interest (a little too broad and general if you ask me). It looks like their current focus is on in vivo imaging. The part that troubles me a little. It’s a lot of the old guard, albeits a ton of heavyweights. I don’t get the mental image of some of the sharpest minds of the world, including some of the younger, more thirsty, scientists, together somewhere in a facility in Kendall Square, dreaming up solutions to some of the problems that are faced across the pharma industry. Also, the investment (around $39 million) by the three big pharma companies is fairly small, so they aren’t taking a huge risk either. Another concern is a lack of clarity on the long term business model. Will pre-competitive data be available as Open Data? How do they plan to commercialize any discoveries? While I might sound a mite cynical, I hope something good comes out of this. Even if only one or two innovations that make a big dent on bringing out better, safer, drugs come out over the next 5-10 years, the company should be considered a success. That said, a constant theme on this blog is precisely the lack of innovative technological platforms. Why has the venture community not given the development of these platforms a chance? Personally, despite the challenges, we need to think about new models to develop such platforms, whether via public/private partnerships or microfunded projects. Enlight seems to be in the former category, so will be interesting to see how it evolves. Further reading: |
| UC Davis Med School's conflict of interest policies among best [The Tree of Life] Posted: 12 Jul 2008 11:20 AM CDT A little late on tis post but still wanted to point out this story in the Davis student newspaper (California Aggie - UC Davis Med School's conflict of interest policies among best). They report that: The American Medical Student Association recently conducted a ranking of medical schools based on their policies regarding free gifts from pharmaceutical companies. UC Davis was one of only seven schools nationwide who received a grade of "A" - meaning the school has a comprehensive policy that restricts pharmaceutical company representatives' access to both campuses and academic medical centers.Good to see this. I find the layers of real and potential conflicts of interest in medical research in general to be very poorly handled by the community. Not that basic science is immune to this problem either but it seems worse in medical research. For some scathing commentary on conflict of interest in medical research, keep an eye on Steven Salzberg's blog. Every once in a while he has some juicy stuff to discuss. |
| Epigenetics and Neo-(Neo-)Lamarckism. [T Ryan Gregory's column] Posted: 12 Jul 2008 06:28 AM CDT A very brief comment on a complicated topic... |
| SNPedia, bioinformatics and Frank Zappa in Vilnius [www.cancer-genetics.com] Posted: 12 Jul 2008 02:18 AM CDT This week I was really happy to meet the founder of SNPedia Mike Cariaso. And not virtually, but in reality in my native city Vilnius, where a bioinformatics EuroPhyton conference was held. Some walk in the old town and beer was great. And there was even famous Hans Rosling talk in this conference! (what a pity, I didn’t manage to listen to it, but it was pretty much the same he had at TED conference:
This website shares information about the effects of variations in your DNA, citing peer-reviewed scientific publications. Great stream from Bethesda, MD, just opposite National Health Institute And there is something unique in Vilnius, a blend of American (unnoficial) culture - the monument of Frank Zappa - the only such kind of monument for rock musician in a world. Zappa's death from prostate cancer in 1993 hit photographer Saulius Paukštys and fellow devotees hard and in a flash of inspiration they managed to erect a lasting memorial to this truly libertarian person.  |
| Posted: 12 Jul 2008 02:16 AM CDT I consider myself, as Dean Giustini would say, a “medupunk“. If you haven’t heard about edupunks:
The world of medicine is changing. Be ready for some interesting projects during the summer. Until then:  This posting includes an audio/video/photo media file: Download Now |
| The challenges of psychiatric genetics [Genetic Future] Posted: 12 Jul 2008 01:33 AM CDT  Back in April I posted a long slab of uninterrupted text on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year. Back in April I posted a long slab of uninterrupted text on the elusive genetics of bipolar disorder, a crippling psychiatric condition affecting over 2% of the population in any given year.The major message from that article is that although bipolar disorder is massively influenced by genetic factors (around 85% of the variation in risk is thought to be due to genetics) we still don't really have the faintest idea exactly which genes are involved. This is despite three reasonably large genome-wide association studies involving over 4,000 bipolar patients in total, which generated weak and contradictory results and failed to provide a single compelling candidate for genetic variation underlying this disease. This disappointing result has also held largely true for other psychiatric conditions with strong genetic components, such as schizophrenia, major depression and autism. Genetic studies of these conditions have had some success identifying rare mutations that underlie severe cases, but the vast majority of the genetic variants contributing to risk remain undiscovered. There are several reasons why genome-wide association studies can fail to yield significant harvests of disease-associated genes. I summed these up with respect to bipolar disease as follows: The researchers are surely hoping that small effect sizes are the major problem, since this is the easiest problem to remedy (simply increase sample sizes). Disease heterogeneity - in other words, multiple diseases with distinct causes that all converge on a bipolar end-point - also seems like a particularly plausible explanation given the complexities of mental illness. It's also likely that various types of genetic variants that are largely invisible to existing SNP chips, like rare variants and copy-number variation, are important.The same story probably holds largely true for other psychiatric conditions. In this week's issue of Nature, a news article and an editorial both tackle the challenges of psychiatric genetics, and lay out the ambitious strategies currently being pursued by researchers around the world to overcome them. Small effect sizes The first hurdle that I describe above is the fact that most of the variants underlying these conditions probably have very small effect sizes (only increasing risk by less than 20%). Such variants will only be identified by cranking up sample sizes immensely, an approach that has yielded some limited success for other genetically complex traits such as height and obesity. The Nature news feature has a table listing some of the major collaborative efforts currently collecting genetic information from the very large cohorts required to dissect out the basis of these conditions: 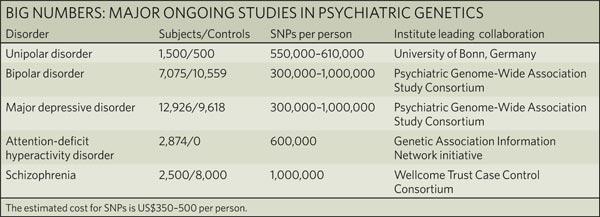 In most cases, these samples are being built up by pooling results from multiple different studies, often gathered by groups from around the world. As sample sizes increase the power of studies to detect small-effect variants grows. The effect of sample size on the power of genome-wide association studies is illustrated in the graph below (which I've managed to save without recording a source - if anyone recognises it, please let me know): 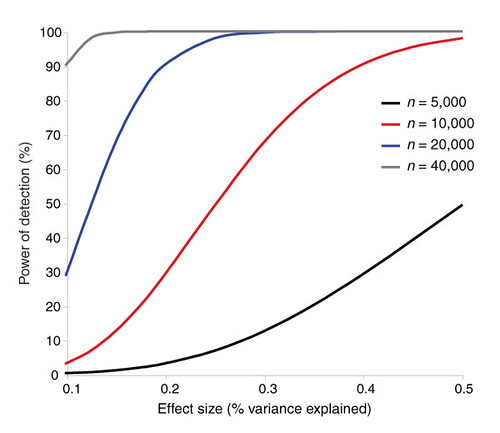 Take a single genetic variant that explains just 0.5% of the variance in the risk of a psychiatric disorder. With a sample size of 5,000 individuals with that disorder you still have a mere 50% chance of detecting that variant. Double your sample size, and that probability jumps to a near-certainty of detection - and your power of detecting even smaller-effect variants (explaining, say, 0.2% of the risk) starts to climb to respectable levels. By staring at those curves for a while, and bearing in mind that many of the variants found by recent genome-wide association studies explain well under 0.2% of the risk variance, you will quickly start to appreciate why researchers are pushing for ever-larger disease populations to work with. With truly enormous samples on the order of 50 to 100 thousand patients - not out of the question for international consortiums studying reasonably common diseases such as bipolar - the power to detect even very weak risk variants becomes reasonable. If there are common genetic variants contributing to the risk of these diseases, such large collaborative studies will eventually find them; so long, of course, as they can tackle the next (and potentially far more serious) problem of disease heterogeneity. Complex, heterogeneous diseases  The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible. The second major problem I mentioned with analysing the genetic basis of these diseases is that they are complex, multifactorial, and extremely difficult to diagnose and classify. Psychiatric conditions are probably the most difficult area of medicine to draw hard boundaries: many symptoms are shared by multiple conditions, and many patients display a diffuse constellation of clinical signs that makes a clean diagnosis impossible.This complexity and heterogeneity is the basis of considerable tension between geneticists and neuroscientists, which is explored in the Nature editorial. Basically, to build up those massive sample sizes shown above geneticists are forced to lump together patients with a variety of clinical symptoms, thus essentially ignoring the complexity inherent in these conditions - a failure that neuroscientists find inexcusable. In turn, geneticists (like myself) get seriously annoyed by the tendency of neuroscientists to make big, bold claims about disease mechanisms based on studies with tiny sample sizes. Both sides make reasonable criticisms. As I said in the quote from my previous article above, I suspect that disease heterogeneity - that is, multiple diseases states with the same broad end point being simplistically lumped together - plays a major role in the failure of genome-wide association studies of psychiatric conditions; at the same time, I find the scientific value of much of the "sexy" neurobiology currently being published (e.g. functional MRI finds that conservatives have lower activity in "compassion" centres of the brain, or whatever) to be highly questionable. Both sides of this scuffle have something to learn from their opponents. The editorial argues, sensibly, that geneticists and neuroscientists just need to start getting along (perhaps a drum circle would help?). The ideal situation is one in which rigorous clinical assessments are used to generate patient cohorts that are as homogeneous as possible that can then be subjected to large-scale genetic analysis. One especially promising avenue is the use of "endophenotypes" - that is, simple and easily quantifiable traits that are sometimes but not always associated with a particular disease. Cleanly defined endophenotypes, such as very specific dysfunctions of brain activity, may prove much more amenable to genetic dissection than the larger, more complex diseases they are associated with. Comprehensively tackling the genetic of psychiatric conditions will require a forceful and combined approach drawing on the clinical expertise of neuropsychiatrists and the experience of geneticists in unravelling the genetic mechanisms of complex traits. To some extent this is happening already (no large genetics consortium would be naive enough to embark on a multi-million dollar project without consulting clinical experts) - but obviously there is considerable room for improvement. Moving beyond common SNPs  Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation. Current genome-wide association studies currently rely largely on the use of single-letter variations in DNA called single nucleotide polymorphisms (SNPs), mainly because these are easy to analyse and can be simultaneously analysed in their hundreds of thousands using chip-based assays. For various reasons almost all of the SNPs on current genome-wide association chips are common sites of variation, present at a frequency of 5% or more in the population. However, recent studies have made it look increasingly likely that a large proportion of the genetic risk of common diseases lies in types of genetic variation that cannot be detected using common SNPs: rare variants, and large-scale rearrangements of DNA known as structural variation.The approaches required to capture these variants are already pretty well-known, although they remain expensive and technically challenging. In an ideal world, genome-wide association studies would be truly genome-wide - in other words, they would utilise the entire DNA sequence of all of the patients and controls in the sample to find every possible genetic variant that might contribute to disease. Unfortunately, such an approach is currently out of reach, for several reasons:
Both approaches have their limitations. The success of the candidate gene approach will be constrained by researchers' ability to identify the genes most likely to be involved in a particular disease - but in fact our currently severaly limited understanding of disease genetics is precisely why we need to study this issue in the first place! (In the Nature news piece, Harvard's Steven Hyman memorably describes this approach as "like packing your own lunch box and then looking in the box to see what's in it.") And while chip-based detection of structural variation is rapidly increasing in resolution, it's extremely difficult to determine which of the variants identified in a study are disease-causing and which are harmless polymorphisms - this is currently done probabilistically, by showing that there is an enrichment of new variants in disease cases compared to controls, but this approach cannot tell you which of the identified variants are actually causative. From psychiatric genetics to genetic psychiatry? There are several important reasons researchers are interested in the genetics of mental illness: identifying causal genes helps to dissect out the molecular pathways involved in disease, and may help to pull out otherwise invisible sub-types of a disease; studying "extreme" mental phenotypes may illuminate the genetic basis of variation in cognition and personality traits in "normal" people; and, perhaps most importantly, by identifying the genes underlying psychiatric diseases we may be able to target at-risk individuals for monitoring and intervention, potentially heading off severe disease before it takes hold. In the headlong pursuit of these goals the field of psychiatric genetics has developed an unfortunate reputation built on bold claims made with limited evidence, and literally hundreds of reported associations that have completely failed to stand up to replication. Just a couple of years ago the shiny new tools of large-scale genomics promised an end to this ignoble period in the history of the field; unfortunately, the introduction of larger samples, higher genomic coverage and increased statistical rigour has not brought the desired clarity to the field, but rather seems to have increased the levels of confusion and uncertainty. If anything, that crucial third goal - using genetic to predict the risk of mental illness - now appears further away than it did just a couple of years ago. Back in early 2007 we didn't have many convincing genetic predictors of mental illness, but at least it was possible to imagine that emerging genomic technologies might identify a small core set of large-effect variants that would help clinicians to predict disease risk. Right now we still don't have many useful genetic predictors, and that illusion of hope is gone. In summary: while there's no doubt that these conditions do have a strong genetic basis, it's now abundantly clear that this basis is frighteningly complex, with common variants of moderate-to-large effect - the types of variants that would be most useful for risk prediction - being essentially absent. It's going to take many years, massive cohorts, the clever application of new genomic technologies, and a willingness from both neuroscientists and geneticists to listen to one another to move this field forward. (Brain scan image from Science Photo Library.) |
| Predicting responses to genetic testing [Genetic Future] Posted: 12 Jul 2008 01:30 AM CDT In a letter published in Nature last week, Kenneth Kosik and Francisco Lopera argue, based on their experiences with a large Colombian family with high rates of Alzheimer's disease, that responses to genetic risk information can vary hugely between individuals. Kosik and Lopera criticise a recent Nature news article that questioned whether genetic test results actually have any lasting negative impact on recipients, noting that one of the members of the Alzheimer's family they study said he would "shoot himself in the head" if he discovered he carried the same mutation that had afflicted his mother with early dementia. Arguing with anecdotes is a fun game; perhaps next week I can get a letter in Nature questioning the tobacco-lung cancer connection using the longevity of my chain-smoking great-grandfather as evidence. However, what really interested me was this paragraph, which lovingly describes the dawning of a new era of techno-paternalism: Seeking predictive genetic testing can be a risky behaviour, and an individual's likely response to genetic risk is hard to foretell. Functional magnetic resonance imaging activity patterns may be able to define people who are more comfortable with risk, and genetic polymorphisms seem to contribute to risk-taking behaviour. Defining the scientific basis for how individuals handle volatile genetic information may help guide our decisions about the best setting for delivering predictive-testing news.So if a person tests positive for a genetic variant that indicates they'll respond badly to genetic testing results, should you tell them? |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |



 If you do not know what is
If you do not know what is 

No comments:
Post a Comment