The DNA Network |
| Posted: 30 Jul 2008 07:00 PM CDT Well, I have been out sick for a while. But I am now finally apparently getting better. Thanks to the work of scientists who have developed multiple classes of antibiotics. Anyway, more on that later. While I was out sick I spent a lot of time searching the web for information about nasty, cellulitis causing antibiotic resistant bacterial infections. And I was researching some other health related issues that may have contributed to my getting such an infection. Here are some tidbits I learned during this forced homecation.
Anyway, enough about me. This is just a quick post to say the Tree of Life will be coming back over the next week or two. I am still out sick. But a clear sign to me that I am getting better is that I finally want to blog again. |
| DNA sequencing and bioinformatics in the classroom: an iFinch case study [FinchTalk] Posted: 30 Jul 2008 03:39 PM CDT |
| World Health Organization: 3 Days in Geneva [ScienceRoll] Posted: 30 Jul 2008 11:21 AM CDT About a year ago, I found an article mentioning WHO’s Wikipedia-based approach in revisioning ICD (international classification of diseases) and contacted them by e-mail. Later, some weeks ago, I was invited for a brainstorming to the centre of World Health Organization in the beautiful city of Geneva. I spent three days there and discussed how a wiki-like system could help making this process (the revision of ICD) more open and collaborative. The centre of World Health Organization Inside WHO Me on the top of WHO with the lake in the background Le jet d’eau is really spectacular (140 metres high). United Nations I guess PubMed users come together for a drink in ClubMed… So those people working in WHO do a huge job. The ICD codes are the basic elements of any kind of health statistics and global healthcare-related decisions are based on these. If this revision process becomes open for all the physicians in the world, it will be even more efficient. The first step of this long process is here. That’s why a wiki-like system could be beneficial and that’s why they are really open to ideas and thoughts. I hope they could use my experience or knowledge or whatever I have in this field of medicine. From now I will keep you posted about how WHO is using the advantages of web 2.0.  |
| All people have time bombs! [The Gene Sherpa: Personalized Medicine and You] Posted: 30 Jul 2008 09:56 AM CDT |
| Posted: 30 Jul 2008 08:38 AM CDT |
| wellcome to DNA network [Reportergene] Posted: 30 Jul 2008 07:37 AM CDT  In order to better reach such hidden niche of passionates, Reportergene will proudly broadcast its specialized view in reportergenomics to the DNA network, a group of up to 50 entusiast bloggers joined together in a feedburner network. If you're looking for information regarding DNA, genes, genomes and (of course) any reporter gene, subscribe the DNA Network RSS feed that aggregates all these amazing blogs into one power packed feed source. In order to better reach such hidden niche of passionates, Reportergene will proudly broadcast its specialized view in reportergenomics to the DNA network, a group of up to 50 entusiast bloggers joined together in a feedburner network. If you're looking for information regarding DNA, genes, genomes and (of course) any reporter gene, subscribe the DNA Network RSS feed that aggregates all these amazing blogs into one power packed feed source.Read older site news |
| Patients out of patience [genomeboy.com] Posted: 30 Jul 2008 07:28 AM CDT The revolution continues…
|
| Phenotype of the day [genomeboy.com] Posted: 30 Jul 2008 07:26 AM CDT
|
| Therapeutic Alchemist [Sciencebase Science Blog] Posted: 30 Jul 2008 07:00 AM CDT Over on ChemWeb and wearing my Alchemist hat I hear of a discovery that could lead to a new therapeutic target for a whole range of diseases in which the inflammatory response is involved, almost a medical Panacea. An out of this world approach to liquid telescopes could overcome the big obstacle in making such a device useful for astronomy. The protein spike on the surface of the Ebola virus is laid bare by X-ray crystallography and could lead to new treatments for slowing outbreaks. Birds of prey could be the new environmental “canary” when it comes to toxic heavy metal, according to Spanish researchers. Japanese researchers have taken individual rotaxanes for a spin and obtained some dynamic snapshots. Finally, the Michael J Fox Foundation has announced its annual round of funding for Parkinson’s therapeutic lead research. Get the fully skinny and the links… a |
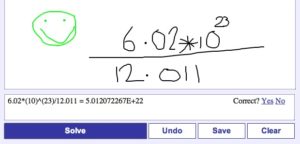
| Draw Your Calculations, xThink does the Math [Bitesize Bio] Posted: 30 Jul 2008 05:38 AM CDT If you use your calculator a lot, especially for complicated equations, and would like a more visual interface then xThink’s Online Calculator could be right up your street. Instead of linearly typing your equation into the calculator, xThink’s revolutionary interface allows you to draw the equation using your mouse (or even better, a digital pen). Hit “solve” and the calculator tells you how it has interpreted your drawing and gives you the solution. You can also annotate (e.g. by adding a green smiley face) and save calculations for later use. See my example below.
The calculator is still in beta testing and initially I found it a bit frustrating as it often misunderstood my drawing. However after reading the instructions (!) it became clear that the problem was mine and not the software’s. For multiplication, you should draw a *, not an “x” and for division, the line should not be shorter than either the numerator or denominator. Also, be sure to not to draw the numbers and symbols too small so that they are as clear as possible. Those three little tidbits solved all of the problems I initially had. This is a great tool that is definitely going on my bookmark list. If you give it a go, tell us what you think here. |
| Posted: 30 Jul 2008 03:11 AM CDT Yesterday, Bioinformatics accepted this paper of mine:
|
| What do people do with health information from their DNA? [Yann Klimentidis' Weblog] Posted: 29 Jul 2008 11:44 PM CDT This project partly aims to determine how people respond to personal genomic information about their health risks. Here's an NIH news story about the study. The money line from the abstract: "We recommend that a considerable research commitment be made now in order to successfully bridge the rapidly widening gap between gene-disease association research and the critical (but slower and more involved) investigations into public health and clinical utility." Putting science over supposition in the arena of personalized genomics Colleen M McBride, Sharon Hensley Alford, Robert J Reid, Eric B Larson, Andreas D Baxevanis & Lawrence C Brody Nature Genetics 40, 939 - 942 (2008) Abstract: We explore the process of going from genome discovery to evaluation of medical impact and discuss emerging challenges faced by the scientific community. The need to confront these challenges is heightened in a climate where unregulated genetic tests are being marketed directly to the general public1, 2. Specifically, we characterize the delicate balance involved in deciding when genomic discoveries such as gene-disease associations are 'ready' to be evaluated as potential tools to improve health. We recommend that a considerable research commitment be made now in order to successfully bridge the rapidly widening gap between gene-disease association research and the critical (but slower and more involved) investigations into public health and clinical utility. Lastly, we describe a large, ongoing, early-phase research project, the Multiplex Initiative, which is examining issues related to the utility of genetic susceptibility testing for common health conditions. |
| The youngest DNA author? [Omics! Omics!] Posted: 29 Jul 2008 09:45 PM CDT Earlier this year an interesting opportunity presented itself at the DNA foundry where I am employed. For an internal project we needed to design 4 stuffers. Stuffers are the stuff of creative opportunity! A stuffer is a segment of DNA whose only purpose is to take up space. Most commonly, some sort of vector is to be prepared by digesting with two restriction enzymes and the correct piece then purified by gel electrophoretic separation and then manual cutting from the gel. If you really need a double digestion then the stuffer is important so that single digestion products are resolvable from the desired product; the size of the stuffer causes single digests to run at a discernibly different position. Now, we could have made all 4 stuffers nearly the same, but there wasn't any significant cost advantage and where's the fun in that? We did need to make sure this particular stuffer contained stop codons guarding its frontiers (to prevent any expression of or through the stuffer), that it possess the key restriction sites and that it lack a host of other sites possibly used in manipulating the vector. It also needed to be easily synthesizable and verified by Sanger sequencing -- no runs of 100 As for example. But beyond that, it really didn't matter what went in. So I whipped together some code to translate short messages written in the amino acid code (obeying the restriction site constraints) and wrap that message into the scaffold framework. And I started cooking up messages or words to embed. One stuffer contains a fragment of my post last year which obeyed the amino acid code (the first blog-in-DNA?); another celebrates the "Dark Lady of DNA". Yet another has the beginning of the Gettysburg Address, with 'illegal' letters just dropped. Some other candidates were considered and parked for future use: The opening phrase to a great work of literature ("That Sam I am, That Sam I am" -- the title also work!), a paen to my wagging companion,. But the real excitement came when I realized I could subcontract the work out. My code did all the hard work, and another layer of code by someone else would apply another round of checks. The stuffer would never leave the lab, so there was no real safety concern. So I offered the challenge to The Next Generation and he accepted. He quickly adapted to the 'drop illegal letters' strategy and wrote his own short ode to his favorite cartoon character, a certain glum tentacled cashier. I would have let him do more, but creative writing's not really his preferred activity & the novelty wore off. But, his one design was captured and was soon spun into oligonucleotides, which were in turn woven into the final construct. So, at the tender age of 8 and a few months the fruit of my chromosomes has inscribed a message in nucleotides. For a moment, I will claim he is the youngest to do so. Of course, making such a claim publicly is the sure recipe to destroying it, as either someone will come forward with a tale of their toddler flipping the valves on their DNA synthesizer or will just be inspired to have their offspring design a genome (we didn't have the budget!). And yes, at some future date we'll sit down and discuss the ethics of the whole affair. How his father made sure that his DNA would be inert (my son, the pseudogene engineer!) and what would need to be considered if this DNA were to be contemplated for environmental release. We might even get into even stickier topics, such as the propriety of wheedling your child to provide free consulting work! |
| The accelerated world of molecular simulation [business|bytes|genes|molecules] Posted: 28 Jul 2008 11:05 PM CDT
For the first time since MM-PBSA came around and people started applying MD to study large scale molecular motions (just see work by Benoit Roux and Georgios Archontis), I am really encouraged about the state of molecular simulation. GPUs, acceleration and large scale distributed computing, coupled with special purpose machines are poised to give the field a short in the arm. I have always been annoyed by the fragmentation in the field, but I really like what I’m seeing over the past few months. To focus on building good engines, writing good software and emphasizing usability and performance. I still maintain that molecular simulation is the scientific field that I have enjoyed the most, but the lack of innovation had made me a little cynical. While I don’t see the impact on drug discovery/design quite yet, but I think we are close to that point in time. Will be watching the space like a hawk. Disclaimer: My day job is very much all about large-scale distributed computing Related articles by Zemanta
|
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |









No comments:
Post a Comment