The DNA Network |
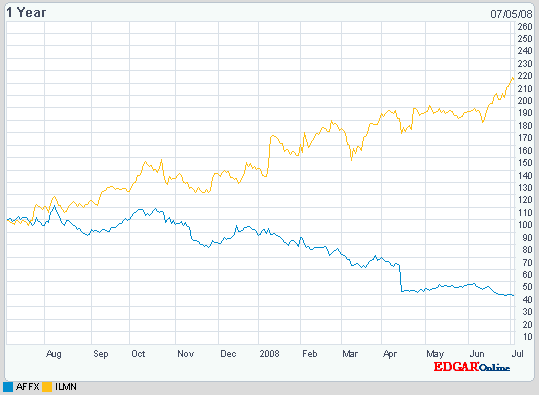
| Good times for Illumina [Genetic Future] Posted: 23 Jul 2008 06:23 PM CDT Genetic analysis company Illumina has just reported a very good quarter: a 66% increase in both revenue and profits. What are they doing right? I've posted a number of times recently about the wave of genome-wide association studies (GWAS) that has surged through the field of human genetics over the last two years. Put simply, GWAS involve looking at up to a million sites of common genetic variation throughout the genomes of thousands of disease patients (cases) and healthy people (controls), and identifying variants that are more common in cases than controls. Although I've tended to focus somewhat on the failures of the GWAS approach (see the notable examples of height and bipolar disease, and a list of the major pitfalls of GWAS), it's worth noting that this approach has nonetheless yielded more information about the genetic basis of common diseases and complex traits in the last two years than we learnt from decades of previous research. A recent review of the field listed nearly 100 genetic variants that have been reliably associated with risk for a total of almost 40 different common diseases or traits, compared to a mere handful of well-validated genetic risk factors prior to the GWAS era - and those numbers are already seriously out of date, with new GWAS being published at an amazing rate.  The technological magic that has enabled the GWAS explosion is the development of genotyping chips: tiny glass chips covered with probes, which - when bound to fluorescently tagged DNA - provide extremely accurate read-outs of the sequence at hundreds of thousands of common variable sites (called SNPs). The picture on the left shows the Illumina Human 1M-Duo chip, which can simultaneously analyse over one million SNPs from two different samples in a single run. The technological magic that has enabled the GWAS explosion is the development of genotyping chips: tiny glass chips covered with probes, which - when bound to fluorescently tagged DNA - provide extremely accurate read-outs of the sequence at hundreds of thousands of common variable sites (called SNPs). The picture on the left shows the Illumina Human 1M-Duo chip, which can simultaneously analyse over one million SNPs from two different samples in a single run.The two major rival chip-makers on the market for quite some time have been Affymetrix and Illumina, both of whom produce chips to analyse genetic variation and also to look at gene expression levels (using a technique called microarray analysis). You don't need to be a financial expert (luckily, because I'm not one!) to see that Illumina is doing something better than Affymetrix: over the last twelve months Illumina shares have more than doubled (and are currently at an all-time high), while those of its rival have halved over the same period:
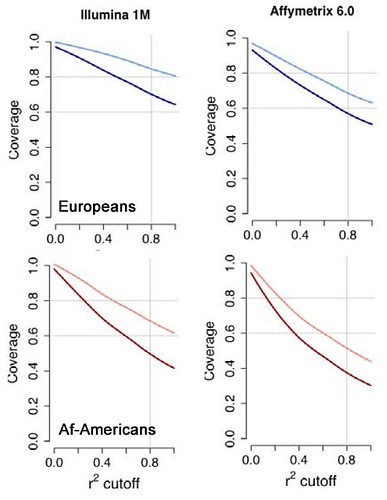
As I said, I'm unqualified to comment on the financial aspects of this increasingly one-sided race, but there are a few scientific issues that distinguish between the two companies. Firstly, when it comes to genome-wide SNP chips, Illumina seems to offer a superior product, at least according to an interesting paper in Nature Genetics a couple of weeks ago. The paper, from a group at the University of Washington, set out to evaluate the performance of large-scale genotyping data (the type used in GWAS) to capture the totality of human genetic variation. They did this using full sequencing data from 76 genes in individuals from various human populations, generated as part of the ongoing SeattleSNPs project, which they could then use as a test set to determine the true coverage of each of the competing genotyping platforms. Illumina came out of the analysis a clear winner. In the image below I've reformatted data from two separate figures from the supplementary data of the paper to make the comparison easier:
There's a lot of data in those graphs, but I want to use them to illustrate two points: (1) for both populations the line for the Illumina 1M chip falls more gradually than that for the Affymetrix chip, indicating that the Illumina chip provides better coverage overall; and (2) this trend is even more marked in the African-American samples, in whom the genetic heterogeneity of their African ancestors makes SNP-based analysis more challenging. The extra coverage provided by the Illumina 1M chip was presumably a factor in the announcement a few weeks ago that the Wellcome Trust Case Control Consortium (WTCCC) - the largest GWAS consortium in the world - will be using this chip for at least part of its planned analysis of over 90,000 common disease patients, which I wrote about back in April. That sort of order must have put a smile on the faces of Illumina executives. Increased sales of genotyping chips for GWAS applications have played a substantial role in the companies' success, according to GenomeWeb News. However, while genome-wide genotyping is currently a major feature of large-scale human genetic research, it's also a technology with a limited life expectancy: in essence, it's just a temporary place-holder for the moment when whole-genome sequencing becomes feasible. Over the next few years we will no doubt see chips with increasingly large numbers of markers crammed in, including rare variants, small insertions and deletions, and structural variation - but no matter how good these chips get they will be rendered completely obsolete by cheap sequencing technology. Why be content with assaying one or two or even ten million markers, when you can have the sequence at every position in your genome for a few thousand dollars? Cheap sequencing also threatens the other major application of chip technology, microarray analysis of gene expression: using a new approach called RNA-Seq, researchers can now effectively use large-scale sequencing technology to read out which genes are being expressed in a tissue, and at what levels. The advantages of this technology over traditional chip-based microarray are considerable: the approach provides information on essentially every gene expressed in the cell (while microarray is limited by whether or not a probe exists on the chip for that gene); it is able to detect exactly which version (alternative splice form) is being expressed; and it has a much higher dynamic range for expression levels, which is limited for chips by the binding capacity of each probe. A couple of weeks ago we saw an application of RNA-Seq to human cells (reported in GenomeWeb News): in an analysis of gene expression in just two cell lines, the authors identified a swathe of novel expressed regions of the genome, and over 4,000 previously unknown positions of alternative splicing (a mechanism used by genes to make multiple proteins from the same DNA sequence). It's clear from the power of this technology that the days of chip-based microarrays are numbered.  Luckily, Illumina has a backup plan. Even as its chip business has grown, Illumina has been fostering the very technology that will make its own chips redundant: next-generation sequencing. Illumina's range of next-gen sequencing machines, the Genome Analyzer series (left), represent one of the big three platforms currently competing furiously for the large-scale sequencing market. In addition, this week the company announced the acquisition of a second sequencing technology, a long-read pyrosequencing platform developed by Avantome. Luckily, Illumina has a backup plan. Even as its chip business has grown, Illumina has been fostering the very technology that will make its own chips redundant: next-generation sequencing. Illumina's range of next-gen sequencing machines, the Genome Analyzer series (left), represent one of the big three platforms currently competing furiously for the large-scale sequencing market. In addition, this week the company announced the acquisition of a second sequencing technology, a long-read pyrosequencing platform developed by Avantome.Affymetrix also has a large-scale sequencing technology, using a chip-based approach - but the limited scale and inflexibility of this system means it has been rapidly overshadowed by the more powerful next-gen sequencing platforms. Anyway, it's clear that the genetic analysis industry is currently at an inflection point, with chip-based technology still on the ascendant but visibly doomed by the simultaneous explosion of next-generation sequencing. Illumina seems well-placed to make a successful - and profitable - transition through this chaotic period. Am I being over-optimistic about Illumina's success? Let me know what I'm missing in the comments. |
| Protein Folding… With Playstation 3? [] Posted: 23 Jul 2008 05:34 PM CDT
This is a pretty amazing idea: Donate CPU cycles on your Playstation 3 for Stanford University’s Folding@Home project. How’s that for a compelling argument for buying Sony’s hyper-addictive gaming station? Check out the YouTube video for all the details. Though we’d rather see an interactive protein folding module for Wii. |
| ISMB 2008: micro-blogging at its best [The Seven Stones] Posted: 23 Jul 2008 04:49 PM CDT Probably like many others, I have often been puzzled by the phenomenon of 'micro-blogging', which consists in posting very short messages on the web (typically via sites such as Twitter) with the goal of providing an instantaneous description of the activity, state of mind or thoughts of the writer. The last few days, a small group of bloggers attending the ISMB 2008 Conference in Toronto used a form of collective micro-blogging on FriendFeed in an intensive way to cover many of the talks held at the conference. Particularly interesting was the coverage of several keynote lectures, often commented simultaneously on a single 'feed' by several bloggers in the audience, providing so to say a real-time example of 'crowdsourcing'. The result is a surprisingly useful set of notes, where the combined attention and complementary knowledge of the participants allow some gaps to be filled, provide additional information (including references or links) and follow the flow of the presentation as it unfolds. I provide below a few picks, relevant to systems biology, while the rest can be consulted (and, importantly, searched!) in the ISMB 2008 Room' on FriendFeed. Good job & many thanks! |
| Six Degrees of Scientific Separation [The Daily Transcript] Posted: 23 Jul 2008 04:21 PM CDT I have a favour to ask of all of you. Go and fill in SciLink's Tree of Science (you'll have to sign up to SciLink first). Why? Well it is very interesting to see how different scientists are connected. And on top of that we can settle a longstanding dispute - what is the appropriate Erdos Number for biologists. You might be asking, what is the Erdos Number? or who was Paul Erdos Number? From an old post by RPM: Paul Erdos was an extremely prolific and mobile mathematician who has left a legacy in academia in the form of the Erdos Number -- a count of your "academic distance" from Erdos. Anyone who published a paper with Erdos has an Erdos number of one (Erdos, himself, had a number of zero), people who published with anyone with an Erdos number of one have an Erdos number of two, and so on. It's a point of pride for a mathematician or other researcher to have a small Erdos number. But who exactly? As a cell biologist I would say that George Palade and Keith Porter would be a good start. Another possibility would be Watson and Crick. In any case to figure out the biology equivalent we would need some sort of repository, or even better some navigational network that lets you explore any possible connection. And now we have the perfect application.
I must say that SciLink's Tree of Science is more of a Genealogical tree ... as in "X mentored Y" as opposed to "X coauthored a paper with Y". In fact some connections such as "coworkers" may not be represented in coauthorships. For example, I never published a paper with my coworker Bil, but we are connected in this tree by the fact that we were in the Rapoport lab at the same time. The opposite problem also occurs. For example, I coauthored a paper with Rich Vallee but according to the current version of the tree we are 4 degrees apart. Of course I could link us up directly ... Here are some other interesting connections. I am Let's take advantage of this user generated content filled application. Plus, it's a great way to kill time in between those long time points. Read the comments on this post... |
| Calling for a moratorium… [genomeboy.com] Posted: 23 Jul 2008 04:20 PM CDT …on George Church stories. I forced myself not to read this until I got my hard copy of Wired. I am sorry to report that it’s really, really good. Please don’t write a book in the next year, Thomas, or I’ll have to have you killed. |
| Sandystroll at Dick Dawkins.net [Bayblab] Posted: 23 Jul 2008 03:14 PM CDT A very excellent series of posts at Sandystroll on good science writing has been posted at richarddawkins.net. Larry has a bit of a beef with some of Sir Dawkin's metaphors. He has some good points, and Sandywalk has more consistently relevant comments than richarddawkins.net. |
| Phenotype of the day [genomeboy.com] Posted: 23 Jul 2008 10:56 AM CDT What could be more adaptive than this?
I used to hate to cook. Now I find it gives me immense pleasure, though my daughters are not always enamored of my efforts. For example, I think my fusion chicken is to die for. But you have to like cilantro and some folks are not genomically wired that way. You know who you are. |
| Batman's Echolocation [Bayblab] Posted: 23 Jul 2008 10:32 AM CDT  In a bit of a stretch in an otherwise amazing film, Batman: The Dark Knight, the caped crusader uses cellphone signals to enable his own echolocation technology. It was fitting since bats use echolocation for navigation and prey location. I don't have any idea how feasible something like this would be although I would suspect highly impossible. I just found some interesting things about echolocation. FoxP2 is a gene that the bayblab has discussed previously, and has sequence variation and patterns of expression linked to human speech defects, the evolution of language, and vocal learning in animals. An interesting paper shows that bats have a great variation in the FoxP2 and the authors speculate this is due to selection for echolocation. Also found some information that humans are capable of limited echolocation. Here is a older story about Ben Underwood who from what I have read is basically a bat. Here is a 'documentary' on Ben. |
| Dent, I've found you! [The Daily Transcript] Posted: 23 Jul 2008 08:50 AM CDT If you are a postdoc or a junior faculty in the biomedical sciences, you have probably seen this infamous cartoon by Dent:
Yesterday I was talking with my good pal Dan and we were discussing a few other great pieces by Dent. After that conversation, I did what anyone else would do, I Googled the guy. Little did I know, I had just stumbled into a gold mine! Here it is: http://dentcartoons.blogspot.com/ Proceed with caution. Read the comments on this post... |
| New white paper on our updated assembly algorithm [Next Generation Sequencing] Posted: 23 Jul 2008 08:40 AM CDT We have just released a scientific white paper which confirms, that, in benchmarking tests, our new algorithm for assembly of Next Generation Sequencing data is indeed fast. Not only is our algorithm fast, but it also provides a better quality of the results, compared to other algorithms benchmarked in the white paper. Assistant Professor at Rutgers [...] |
| Architect for innovation [business|bytes|genes|molecules] Posted: 23 Jul 2008 08:13 AM CDT
He said this in the context of building a platform that allowed others to be innovative, e.g. Firefox and it’s plugin architecture. I immediately texted myself that phrase. As life scientists, we really lack such platforms. One reason I liked Pipeline Pilot when I saw it was because, it was like a drawing board for ideas. Cytoscape has that potential (although could be easier to use), as does Taverna, although it’s a little heavy too. Bioclipse probably falls into that category as well. Next time a company, or a developer goes about developing a software suite for molecular modeling, or expression analysis or whatever, keep that phrase in mind, architect for innovation |
| Personalized Medical Search Engine: New Databases [ScienceRoll] Posted: 23 Jul 2008 07:51 AM CDT Scienceroll Search is a personalized medical search engine powered by Polymeta.com. You can choose which databases to search in and which one to exclude from your list. It works with well-known medical search engines and databases and we're totally open to add new ones or remove those you don't really like. Now we updated the list of databases and created some new boxes as well. So we have a Basic Health Information box for those non-experts who would like to find relevant health-related information; and a drug box for drug-related searches only. We also havea box for organizations (e.g.: if you’re interested in the data of WHO or CDC); and another one for Research Information for scientists and medical professionals. Give it a try and, of course, suggestions are welcome! More information:
 |
| Regulatory Placebo [Sciencebase Science Blog] Posted: 23 Jul 2008 07:00 AM CDT
Eric W a chemistry teacher from Minnesota who goes by the online monicker of “Chemgeek” pointed out that the US Food & Drug Administration (FDA) had attempted to do this several years ago (it’s something that has been mooted in the European Union too), but in the US at least all that happened is that products now have a warning such as: "These statements have not been evaluated by the FDA…" Which is obviously a rather weak stand to take on products that can be potentially lethal in combination with the wrong disease or other medication. The problem as I see it, is that if any given herbal product has potent physiological activity, then it is too all intents and purposes, a medicinal drug, and should be tested and labelled as such so that consumers can be warned of contraindications. If the herbal product has no physiological activity, other than perhaps to provide some spurious antioxidants for which our needs are not known, then it is little more than a placebo. Stef Levolger, aka Slevi, is a Dutch medical student with an interesting observation on the issue. “It’s not like [herbal is] all that different from our own basic medicine. Take for example willow bark, sold as a herbal treatment and advertised with [various] uses by the Chinese, by [several] companies and others since the ancient Greeks. And, how do you get it home? Right, you get a bottle filled with pills. I wouldn’t be surprised if it was even produced in the same pharmaceutical factory as where regular aspirin comes rolling down the line, just got a price tag twice as high slammed on it,” he says. I think Slevi is on to something, certainly a huge number of our so-called modern pharmaceuticals have natural product origins while many so-called natural herbal products are manufactured on neighbouring production lines by a division of the pharma companies. Canadian blogger Mina Isabella Murray of Weird Science strongly disagrees with the idea of bringing all herbal remedies under the pharma umbrella. “That’s not to say I don’t support the notion of thorough research, safety, monitoring and accountability, but to compare the vast majority of herbals to pharmaceutical preparations is misguided and excessive,” she says, “I vote stricter regulations but ones that are separate from current pharma protocol/guidelines.” She suggests that the issue is not black-and-white. “The vast majority of herbal preparations aren’t viewed to treat disease in the way that pharmaceuticals are presented nor are they presented as ‘cure alls’, she says, “Sure, we all But, I’d argue that the efficacy of such a “natural” remedy is dubious at best, but other products, may have genuine effects and so can interfere with the metabolism of certain medications. St John’s Wort is probably a case in point. This product widely used to treat mild depression is contraindicated for anyone with thyroid problems. But, buying St John’s over the counter, off prescription as it were, does not provide the necessary warnings. I asked chemical consultant Hamish Taylor of Shinergise Partners Ltd how he felt about regulation of yet another area of his broader industry. “I think the answer should be YES,” he affirms, “but perhaps a sensible step is part-way i.e. strong warnings on possible side-effects and a cautionary ‘there is no clinical trial which demonstrates absolute efficacy’.” Such an approach would allow the placebo effect of “it’s natural, I believe in it, it does me good” to shine through, which is probably no bad thing. “Feeling good is pretty much proven to make people healthier and certainly happier,” Taylor adds. He says that some degree of labelling would however prevent some of the nastier side-effects that can occur. “By going to this quasi-interim step, it may even encourage manufacturers to undertake proper clinical trials to demonstrate effectiveness and therefore encourage greater use,” Taylor says. Of course, it is not as if the manufacturers have not investigated the potential. “The problem is that if the more popular herbal remedies were indeed 100% effective the drug component would have been isolated and purified by now!” adds Taylor. Steve Bannister, Scientific Director & Principal Consultant at Xcelience, LLC, a drug development company based in Tampa, Florida, suggests that really the question we should be asking is what level of regulatory resources can we afford?what level of regulatory resources can we afford? “Natural products have long provided leads for drug discovery,” Banniester says, “In modern natural-product drug discovery, activity-guided fractionation (often using an isolated receptor) identifies an active molecule and a single molecular entity results from semisynthetic improvement of the phytochemical’s drug properties. The drug-regulatory process includes guidelines for determining safety, efficacy, and quality, as well as for setting acceptance criteria for each.” However, some herbal products may have efficacy as a result of a combination of components, additionally adverse reactions to such a product may be due to a different combination. “To pass through the current drug regulatory process, a product specification, including identification of the specific active components and their requisite levels, along with identification and limits for the unsafe impurities, is needed,” adds Bannister. He further explains that this specification must be defined in terms of safety and efficacy in humans. “This is an extraordinarily difficult undertaking, and if required, would prevent herbal products from reaching approval,” he says, “This, combined with the fact that some herbals really do work, is the principal reason for a different different set of regulations.” One has to consider what is an acceptable level of risk and how it is managed? What risks are associated with adulterated products, what are the adverse reactions, and to what extent does lack of efficacy lead consumers away from different, effective, treatments. As it stands, the fully implemented US Good Manufacturing Practices for herbals are meant to control adulteration but can only do so if there is adequate compliance inspection. Of course, evidence of a significant incidence of adverse reactions is sufficient to lead to a product being banned but again this requires significant surveillance resources. Also, disease-treatment claims can be made only for approved drugs, so consumers may have been led to a particular herbal for their symptoms through hearsay or lifestyle magazine “evidence”. Monitoring of such claims for herbals, such as the neurochemical effects of St John’s Wort also requires surveillance resources. It all costs. Delano Freeberg, Chief Technical Officer at API Purifications Inc, suspects that the devil is in the details. “In special cases, increased regulation may be beneficial,” he says, “I believe public health is not served by increased regulation. In fact, I believe we will benefit from the application of well-understood scientific and medical criteria for the reduction of regulation. The benefit of this approach to the public is the timely availability of lower-cost drugs. I believe this can be accomplished with no effect on safety.” He points out that regulation is not required for extracts. “Many herbal remedies are known to have efficacy, even in a crude extract. The composition of this extract often does not differ much from that of the starting biomass (natural herb). If the herb being used is “GRAS” (generally regarded as safe), there is good scientific foundation for believing the extract will likewise be safe,” adds Freeberg, “An important caveat: “Natural” does not imply “safe.” Hemlock extract (Conium maculatum) is 100% natural but also 100% deadly.” He also suggests that regulation should depend on potencyregulation should depend on potency. “Traditional over the counter pharmaceuticals contain a single active component. Natural remedies usually contain several compounds of known efficacy. Green tea, for instance, contains high concentrations of around a dozen different catechin antioxidants. The traditional pharmaceutical OTC preparation must be held to a higher standard of regulation because of the higher purity and potential potency of the active ingredient,” he says. Regulation should also depend on drug type. “I feel the FDA over-regulates and does not take into consideration scientific findings that provide a basis for reduced regulation,” says Freeberg, “I provide one example. Synthetic statins have moderate to severe side effects in 5% of patients. Cholesterol can be equally reduced by the intake of policosanol (One recent study found no efficacy; however, dosage levels may not have been appropriate), a group of long-chain alcohols from beeswax. Side effects of policosanol were mild - no worse than placebo.” There is certainly a scientific basis to demonstrate that many natural compounds have little adverse physiological effect, even in high-purity forms. This can be determined from structure-activity relationships and historical clinical and toxicological data. Florence Leong, an Investment Director at ATP Capital Pte Ltd, believes the answer lies not in regulation but in consumer education. “Consumers need to be educated on the difference between OTC herbal remedies and pharmaceutical products,” she says, “Not all natural products are safe, the reason why OTC have fewer listed side effects as compared to pharmaceutical is because OTC products are not as thoroughly evaluated. Often the long list of side effects in pharmaceutical drugs frightens the consumer and the cursory listing of side effects gives consumer the false perception of safety.” Unfortunately, she adds, consumer education is a long and tedious process and regulation is consider the efficient ‘quick fix’. She echoes my own earlier sentiment regarding efficacy claims where OTC products are not as robustly proven as pharmaceutical products. “This is an area where regulation should be tightened as many manufacturers of OTC products do try to push the limits by making specific efficacy claims in non-print advertisements. And often they do get away with it,” she says. The active constituents of pharmaceuticals products are consistent, which ensures reliable efficacy and, on for established products, predictable side-effects. In contrast, efficacy of OTC herbal medications can be variable due to natural variations of the chemicals in different batches of raw materials, contamination, and manufacture unscrupulousness. This means herbal products can swing between no effects and no side-effects, via those that work and have their own side-effects to the wildly hazardous batch of mercury-laden dessicated ordure. a |
| Ye Olde Antibiotic Plates [Bitesize Bio] Posted: 23 Jul 2008 05:43 AM CDT You are working late in the lab and you need to do a transformation, but sod it, you don't have any antibiotic plates on hand. So you go on the hunt to see if there are any secret stashes anywhere in the lab (you can find secret stashes in every lab if you look hard enough). And lo and behold, come across the ampicillin plates you poured 4 weeks ago then forgot about. But they are old, so how do you know if they will give adequate selection? Should you use them? Luckily, someone way back in 1970 determined the lifespan of a whole host of antibiotics in plates, and the good news is that they last a lot longer than you probably thought. Plates containing methicillin, erythromycin, cephalothin, tetracycline, chloramphenicol, kanamycin, streptomycin, polymyxin B, or nalidixic acid should be a-ok even if stored for one month at 4degC. Ampicillin was shown to have no loss of activity after 1 week but 10% lower activity after 4 weeks at 4°C, which could result in things like satellite colonies, but should be fine for practical purposes. Other antibiotics with reduced activity after 4 weeks included Penicillin G (23% reduced) and nitrofurantoin (17% reduced). So even ampicillin plates should be fine for 4 weeks - certainly longer than I would have thought - meaning that your late night experiment can be saved by using your stash of old plates after all. |
| How deterministic is neighbour-joining anyway? [Mailund on the Internet] Posted: 23 Jul 2008 04:36 AM CDT Whenever we try to publish an algorithm for speeding up neighbour-joining, at least one reviewer will ask us if we have checked that our new algorithm generates the same tree as the “canonical” neighbour-joining algorithm does. This sounds reasonable, but it isn’t really. The neighbour-joining method is not as deterministic as you’d think. Well, it is a deterministic algorithm in the sense that the same input will always produce the same output, but equivalent input will not necessarily produce the same output. The neighbour-joining algorithmThe neighbour-joining algorithm is phylogenetic inference method that takes as input a matrix of pairwise distances between taxa and outputs a tree connecting the taxa. It is a heuristic for producing “minimum evolution” trees, that is trees where the total branch length is as small as possible (while still, of course, reflecting the distances between the input taxa). A heuristic is needed, since optimising the minimal evolution criteria is computationally intractable, and the neighbour-joining algorithm is a heuristic that strikes a good balance between computational efficiency and inference accuracy. The heuristic is not a “heuristic optimisation algorithm” in the sense that simulated annealing or evolutionary algorithms is. It is not randomised. It is a greedy algorithm, that in each step makes an optimal choice, which will hopefully eventually leads to a good solution (although locally optimial choices are not guaranteed to lead to a global optimal choice). A problem with a greedy algorithm is that local choices can drastically change the final result. Not all greedy algorithms, of course. Some are guaranteed to reach a global optimum and if there is only a single global optimum then the local choices will not affect the final outcome (but if there is more than one global optimum the local choices will also influence the final result). In the neighbour-joining algorithm, the greedy step consists of joining two clusters that minimise a certain criteria. If two or more pairs minimise the criteria, a choice must be made, and that choice will affect the following steps. Equivalent distance matrices does not mean identical inputUnless the choice is resolved randomly, and I’m assuming that it isn’t, then the algorithm is deterministic. It could, for example, always choose the first or last pair minimising the criteria. How this choice should be resolved is not specified in the algorithm, so anything goes, really. If the choice is resolved by picking the first or last pair, then exact form of the input matters. The distance matrix neighbour-joining uses as input gives us all the pair-wise distances between our taxa, but to represent it we need to order the taxa to get row and column numbers. This ordering is completely arbitrary, and any ordering gives us an equivalent distance matrix. Not an identical one, though. If we resolve the local choice in a way that depend on the row and column order, then two equivalent input matrices might produce very different output trees. An experimentTo see how much this matters in practice, I made a small experiment. I took 37 distance matrices based on Pfam sequences, with between 100 and 200 taxa. You can see the matrices here. There are 39 of them, but I had to throw two of them away – AIRC and Acyl_CoA_thio — since they had taxons with the same name, and with that I cannot compare the resulting trees. It’s matrices I created for this paper, but I don’t remember the details… it is not important for the experiment, though.
To see a plot of the result, click on the figure on the left. As you can see, you can get quite different trees out of equivalent distance matrices and on realistic data too. I didn’t check if the differences between the trees are correlated with how much support the splits have in the data. Looking at bootstrap values for the trees might tell us something about that. Anyway, that is not the take home message I want to give here. That is that it doesn’t necessarily make sense to talk the neighbour-joining tree for a distance matrix. Neighbour-joining is less deterministic than you might think! |
| TGG Interview Series VIII - Max Blankfeld [The Genetic Genealogist] Posted: 23 Jul 2008 02:00 AM CDT From the “About” page at Family Tree DNA:
In the following interview, Max discusses his lengthy roots with genetic genealogy, the launch of DNA Traits, and the future of genetic genealogy. TGG: How long have you been actively involved in genetic genealogy, and how did you become interested in the field? Have you undergone genetic genealogy testing yourself? Were you surprised with the results? Did the results help you break through any of your brick walls or solve a family mystery? Max Blankfeld: I was introduced to the concept of genetic genealogy at the very beginning when Bennett was talking about doing the “proof of concept” with Mike Hammer at the University of Arizona, even before the name Family Tree DNA had been registered. This was in 1999, and since I have the habit of not trashing my emails, I still have our exchanges on the subject dated from 1999. I certainly did test, in 2000, and while the results did not surprise me, it helped find and confirm distant relationships, and also gave me very close matches with people that I was not aware of. So here’s my DNA break through: in 1983, when I was still in Brazil, our family received a letter from a Blankfield living in Australia, where he gave us some of his genealogy and asked if we could possibly be related. The problem was that my father passed away in 1981, and never discussed very much his family with me because he was a Holocaust survivor, and both parents and sisters were murdered by the Nazis in 1942. So, I didn’t have any facts to check against that letter. I kept the letter in the drawer. Fast forward to the year 2000, and the start of genetic genealogy. I start looking for Blank(en)f(i)elds to be test. Saul Isseroff, an avid genealogist from England tells me that he’s related to some Blankfields in South Africa, and gives me the name of a female Blankfield. She convinces her father to be tested. High expectations. Results come in and bingo - very close match. I ask for their family tree and guess what - that man from Australia is in that tree! We put together both trees, and it looks like we shared the same great-great-great-great-grandfather! (I must say that I had a previous attempt with another Blankfield that did not show a relationship) TGG: Is there any concern that people with little experience in the area of genetic genealogy will confuse ancestry testing with personal genomics services, especially in light of all the recent negative press about personal genomics? MB: Certainly. That is why our first priority has always been to educate before selling. I remember that the first trade-shows that Bennett and I went, we never took kits to sell, and our entire approach was just to make people aware of genetic genealogy and how they could use it. With time, people started coming to our booth and ask if we brought kits to test them. As a former journalist I can tell you that negative press will always be there, no matter what area of business one is in. Unfortunately, many journalists write about topics that they are not experts, and this leads to negative press or some absurd and totally wrong statements (I have good stories from my times as a foreign correspondent, but I will leave this to a different forum). Educating is the key. Not just the customer, but also the press. We spend hours on the phone with journalists from all over the United States and abroad, in very detailed conversations so that they can understand the subject. In fact, our favorite thing is to educate people about it and we do not measure the time we spend on the phone for this purpose. TGG: You recently launched DNATraits together with Mr. Greenspan. Could you tell us a little bit about the new company and what it offers? MB: This is a field that we were reluctant to get in, as genealogists normally don’t like to mix genealogy and health information. However, we noticed that over the years more and more people approached us on the subject of genetic diseases or inherited conditions. This lead us to form a separate entity for this specific purpose, where we use totally different test kits, absolutely unrelated to Family Tree DNA tests and stored DNA. It is currently offering Mendelian tests for several inherited diseases, and we will be adding more tests every few months. We want to change the paradigm in this field, allowing people to get tested for substantially less than what the current market price is. We want for DNATraits to make a difference in this area, and by being very affordable, allow the widest number of people to get tested so that the quantity of people with inherited diseases can be reduced. TGG: What do you think the future holds for genetic genealogy? MB: It may not grow at the rate that we have seen it growing in the past years, but it will still grow, and also, with the team of scientists that Family Tree DNA has, we will keep seeing additional discoveries, and offering additional tests that can help further one’s genealogical research. And, as we integrate additional features in the future, more tools that embed more traditional genealogy along with the results of DNA testing, Family Tree DNA will continue being positioned as the leader, as the most complete and scientifically accurate company in the market - and of course, with the largest database - which is a key element in this field. TGG: Thank you Max for a great interview!
|
| A “biology” programming language? [Mailund on the Internet] Posted: 23 Jul 2008 01:52 AM CDT According to this press release, some people at Harvard Medical School have developed a new programming language for biological modelling. I guess protein modelling from the description, but I could be wrong.
I’m not sure how much of a domain specific or how much of a general purpose programming language it is. They are comparing it with LISP, so maybe it is a domain specific language with some meta-programming features? I haven’t been able to find the paper yet — of course it isn’t published until today and there might not be any preprints online — but I’m curious about this… |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |








 My recent article on the subject of an
My recent article on the subject of an 
 The eighth edition of the TGG Interview Series is with Max Blankfeld. Max is Vice-President of Marketing and Operations at
The eighth edition of the TGG Interview Series is with Max Blankfeld. Max is Vice-President of Marketing and Operations at
No comments:
Post a Comment