The DNA Network |
| I see PLoS in everything [The Tree of Life] Posted: 15 Jul 2008 04:45 PM CDT |
| Posted: 15 Jul 2008 03:58 PM CDT  I have previously been pleased to announce on Genomicron the release of the first two issues of Volume 1 of Evolution: Education and Outreach. I am equally pleased to point out that Issue 3 is now available free online. I have previously been pleased to announce on Genomicron the release of the first two issues of Volume 1 of Evolution: Education and Outreach. I am equally pleased to point out that Issue 3 is now available free online. As a special treat, I note that editors-in-chief Greg and Niles Eldredge mention Genomicron in their editorial.
Here are the contents of Vol 1, Issue 3 Editorial - Gregory Eldredge and Niles Eldredge Some Thoughts on "Adaptive Peaks," "Dobzhansky's Dilemma"—and How to Think About Evolution - Niles Eldredge The Concept of Co-option: Why Evolution Often Looks Miraculous - Deborah A. McLennan Evolutionary Trends - Some Guy The Importance of Understanding the Nature of Science for Accepting Evolution - Tania Lombrozo, Anastasia Thanukos, and Michael Weisberg Do Students See the "Selection" in Organic Evolution? A Critical Review of the Causal Structure of Student Explanations - Abhijeet Bardapurkar Celebrate Darwin Day, An Event for Education and Outreach in Evolutionary Biology - Rachel M. Goodman What Conceptions do Greek School Students Form about Biological Evolution? - Lucia Prinou, Lia Halkia, and Constantine Skordoulis
|
| Gecko genome size and cell size. [Genomicron] Posted: 15 Jul 2008 03:56 PM CDT One of the many aggravations I encounter when reviewing manuscripts is that some authors greatly overstate the applicability of statistically significant patterns they report. For example, a statistically significant pattern in a small comparison of a few animals may be extrapolated in the discussion to the kingdom at large. Today I was disappointed to see a paper that is soon to come out in Zoology that does the opposite -- i.e. takes a non-significant relationship in a handful of species and pretends that it challenges the importance of broad relationships that have been considered important for decades. The paper in question is: Starostova, Z., L. Kratochvil, and M. Flajshans. 2008. Cell size does not always correspond to genome size: phylogenetic analysis in geckos questions optimal DNA theories of genome size evolution. Zoology, in press. They compared genome size and cell size across 15 geckos and found no correlation. From this, they went on to argue that genome size does not causally influence cell size and that genome size is not under selection due to cell size impacts. First, let me point out that strong, positive correlations between genome size and cell size have been reported within and across all vertebrate classes including reptiles. So, on a broad scale, the relationship is clear.
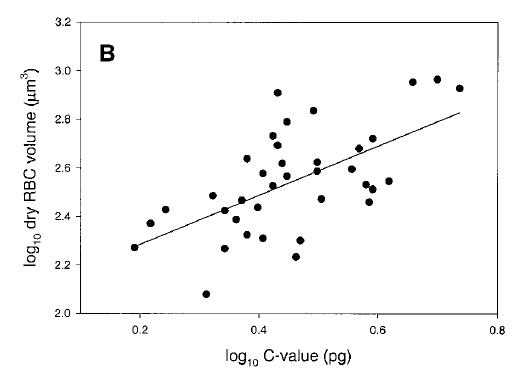
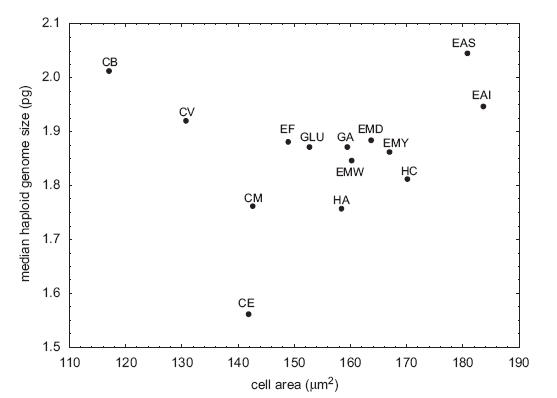
Genome size and cell size in reptiles. From Gregory (2001), based on data from Olmo and Odierna (1982). Second, let me say that I have issues with their methods. For example, they used DAPI as the fluorochrome, which is base-pair specific and can give biased determinations (they recognize this but assume the species are all the same in AT content). Second, they produced fairly substantial error ranges in their measurements given that these were all raised in the lab or obtained from pet shops and not taken from different wild populations (i.e., the variability between conspecifics is probably artifact). Third, they counted "forms" of the same species from different places as being independent in their analyses -- so it wasn't 15 species, rather it was 12 species with several represented by multiple points. These are not the main problems, though. The first is that they clearly had outliers in the dataset. In particular, Coleomyx brevis (CB) and Coleomyx variegatus (CV) have "large" (~2pg) genomes but comparatively small cells. I don't think I even need to draw the line through the remaining points, but in case eyeball statistics don't do it, the correlation is highly significant without them (r = 0.74, p < 0.006) (they recognize this, too, but note the title they chose for the paper nonetheless).
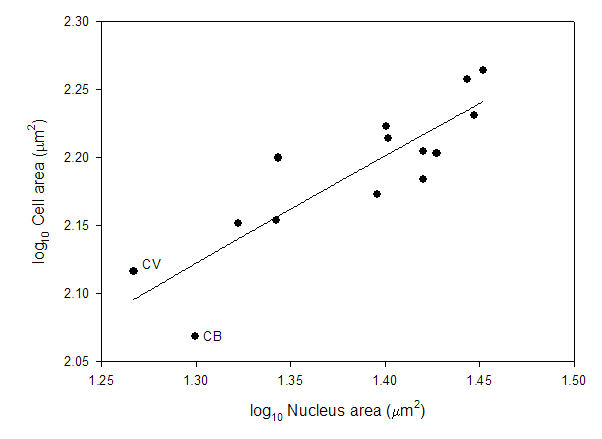
From Starostova et al. (2008). So, how can this be explained? Well, you have to know something about nucleotypic theory, which these authors actually did mention. It's not genome size all alone that is the determining factor -- nucleus size is critical. The "nucleotype" is defined as "that condition of the nucleus that affects the phenotype independently of the informational content of the DNA" (Bennett 1971). As has been pointed out repeatedly (e.g., by me, Cavalier-Smith, Bennett, and others), the compaction level of DNA in the nucleus adds a second dimension to the relationship. More DNA is one thing, but if it is compressed into a tightly packed, reduced nucleus, then cell size may still be small. That leads to the second major problem. Looking at the data reported in a previous study (Starostova et al 2005), there is no correlation between genome size and nucleus size. There is, however, a positive correlation between nucleus size and cell size across these reptiles.
Based on databy Starostova et al. (2005). The two outliers in the genome size vs cell size comparison have more compact nuclei and this allows smaller cell sizes with larger genome size. Cell size is correlated with body size in these geckos, and these two species are "dwarfs" (~4.5g) relative to other species (as big as ~90g). So, there could very easily be selection for reduced cell size which, in this narrow range in DNA amount, was met by a compaction of the nucleus rather than a loss of DNA.This actually reinforces the strength of nucleotypic theory.
______ Bennett, M.D. 1971. The duration of meiosis. Proceedings of the Royal Society of London B 178: 277-299. Gregory, T.R. 2001. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells, Molecules, and Diseases 27: 830-843. Olmo, E. and G. Odierna. 1982. Relationships between DNA content and cell morphometric parameters in reptiles. Basic and Applied Histochemistry 26: 27-34. Starostova, Z., L. Kratchovil, and D. Frynta. 2005. Dwarf and giant geckos from the cellular perspective: the bigger the animal, the bigger its erythrocytes? Functional Ecology 19: 744-749. |
| Abiogenesis vs. evolution. [Genomicron] Posted: 15 Jul 2008 03:54 PM CDT At The Panda's Thumb, Nick Matzke has a post about abiogenesis (the origin of life from non-life) and evolution. He, PZ, and others argue that abiogenesis is part of evolutionary biology and that it is a cop-out to deflect challenges about it from anti-evolutionists. Allow me a brief summary of my interpretation. ---- Question: Do we need evolutionary biology to understand the origin of life? Answer: Very probably. Early replicators, once they arise, would undergo evolution. Mutation, natural selection, etc., would have been important before cellular life as we understand it appeared. However, there are components to the issue that predate the occurrence of natural selection, which are more properly understood in terms of organic chemistry than biology. The line is not sharp, though, so keeping evolution out of abiogenesis research is unwise. Evidence: Whether "cells" (membranes with stuff inside) or replicator molecules (e.g., RNA) appeared first, if there was heritable variation and reproduction and survival that differed among them, then selection would have happened. ---- Question: Do we need to understand how life arose to understand the subsequent evolution of cellular life? Answer: No. Evidence: We have been studying evolution seriously for 150 years and understand quite a bit about how it happens, yet we don't know how life arose. --- So, Nick and others are correct that we should not say that abiogenesis is independent of evolution. However, if they are implying that abiogenesis is part of -- more importantly, a crucial part of -- evolutionary theory, then I do not agree. The influence is one-way, and this is the opposite of the way anti-evolutionists perceive it. They argue that if we do not know how life started, then evolution is false. In actuality, knowing how life started has nothing to do with studying how life has evolved since the first complex cells appeared. However, understanding how complex cellular life evolves probably tells us something about how life arose because the same processes are relevant whenever there are variable replicating entities. In my opinion, it remains a valid and useful argument to point out that uncertainty regarding the origin of life is irrelevant to the factual standing of evolution over the past 3.8 billion years.
|
| Natural selection before Darwin. [Genomicron] Posted: 15 Jul 2008 03:53 PM CDT Charles Darwin (1809-1882) opened his first notebook about "the species question" in 1837, not long after his return from the voyage of the Beagle. By 1838, he had developed the basic outline Some authors have argued that Edward Blyth (1810-1873), an acquaintance of Darwin's, developed the central idea of selection in an 1835 paper in the Magazine of Natural History. For example, Eiseley and Grote (1959) claimed that "the leading tenets of Darwin's work -- the struggle for existence, variation, natural selection and sexual selection are all fully expressed in Blyth's 1835 paper", from which they then quoted the following:
As Eiseley and Grote (1959) and others have noted, however, Blyth's description applies to artificial selection, a process that had obviously been known to breeders long before. In terms of natural selection, Blyth suggested that it would be a conservative and not a creative process, acting to restore the original features modified by artificial selection; that is, it was not a process that caused change, but one that maintained stability within species. Neither Darwin nor Blyth considered this to have been an early example of Darwin's theory of evolution by natural selection (see Schwartz 1974). Patrick Matthew (1790-1874) did not feel the same way as Blyth, and was vocal in his belief that he had preceded Darwin as, according to calling cards he carried, "Discoverer of the Principle of Natural Selection". In fact, Matthew had proposed an idea similar to natural selection in his 1831 book On Naval Timber and Arboriculture,
Darwin, like most others, was not aware of Matthew's book and he learned of it in 1860 when Matthew wrote a letter to the Gardners' Chronicle and Agricultural Gazette including excerpts from his book in response to a review of Darwin's On the Origin of Species. In your Number of March 3d I observe a long quotation from the Times, stating that Mr. Darwin "professes to have discovered the existence and modus operandi of the natural law of selection," that is, "the power in nature which takes the place of man and performs a selection, sua sponte," in organic life. This discovery recently published as "the results of 20 years' investigation and reflection" by Mr. Darwin turns out to be what I published very fully and brought to apply practically to forestry in my work "Naval Timber and Arboriculture," published as far back as January 1, 1831, by Adam & Charles Black, Edinburgh, and Longman & Co., London, and reviewed in numerous periodicals, so as to have full publicity in the "Metropolitan Magazine," the "Quarterly Review," the "Gardeners' Magazine," by Loudon, who spoke of it as the book, and repeatedly in the "United Service Magazine" for 1831, &c. The following is an extract from this volume, which clearly proves a prior claim. In a reply in the same magazine, Darwin said,
In terms of arguments that Matthew's contribution diminishes that of Darwin, I tend to agree with Peter Bowler, who said in his book Evolution: The History of an Idea,
In any case, neither Blyth nor Matthew was the first to propose natural selection in very basic form. Another individual, an American physician of Scottish descent by the name of William Charles Wells (1757-1817), presented a paper in 1813 (published in 1818) that included a process of natural selection to account for differences in skin colour among people. Wells considered light skin to be the primitive condition, with dark skin a subsequent specialization.
Darwin added an acknowledgment of Wells's work beginning with the 4th edition of the Origin in 1866,
What Darwin (and until recently, most historians) did not realize was that the general notion of selection had already been proposed by 1794. In an review in Nature in 2003, Paul Pearson pointed out that James Hutton (1726-1797), considered the Father of Modern Geology, postulated something similar to natural selection in An Investigation of the Principles of Knowledge. (Pearson also noted the interesting bit of trivia that Hutton, Wells, Matthew, and Darwin all attended the University of Edinburgh). As Hutton (1794) said,
Hutton used the examples of fast running and a strong sense of smell in dogs to illustrate the mechanism. He did not, however, accept the notion that new species could form by this or any process. For Hutton, this was limited to creating varieties only and not species.
It is important to note that Darwin himself recognized that others had discovered natural selection at least in basic outline before (and after) he had. He did not avoid sharing credit to the extent that it was due. In light of this history, one could argue that because natural selection as a mechanism had been proposed by several authors that it would have been discovered and recognized as important eventually, even without Darwin's input -- and, indeed, it probably would have, as would Newton's laws of motion, Einstein's theory of relativity, and other fundamental principles describing the natural world. On the other hand, the idea had been around for at least six decades before Darwin published the Origin, and it was not until someone of Darwin's genius developed the idea that evolution assumed its position as the underlying theme of all biology. _______ If you enjoyed this post, please consider subscribing to Genomicron feed.
|
| Epigenetics and Neo-(Neo-)Lamarckism. [Genomicron] Posted: 15 Jul 2008 03:48 PM CDT A very brief comment on a complicated topic... New Scientist has a story in the current issue about epigenetics -- differences in gene expression that are not due to changes in the gene sequences themselves -- and how non-genetic variation can be both influenced environmentally and, in some cases, inherited. The New Scientist story, which is entitled Rewriting Darwin: the new non-genetic inheritance, is another example of the "reporting on a revolution" and "underdog vindicated" fallacies so common in science reporting.
The article includes an interview with Eva Jablonka, who is one of the Altenberg 16 who are, as we speak, revolutionizing evolutionary theory (just kidding, but see here for a summary of what she had to discuss). So, is this "neo-Lamarckism" and is it going to require "a radical rewrite of evolutionary theory"? Some thoughts: 1) There already was a neo-Lamarckism in the late 1800s and early 1900s. So this would be neo-neo-Lamarckism if anything. 2) Epigenetics, by definition, involves modifications of the expression of genetic systems. If heritable, they would be subject to natural selection, drift, etc., when they arise within a population. So, while this is certainly interesting, it will be a welcome expansion of existing theory rather than a revolution. 3) The inheritance of acquired characters was not original to Lamarck (it was the predominant view in his era), and in any case this by itself does not make a theory of evolution "Lamarckian". Lamarckian evolution a) considers adaptation as the result of use and disuse in response to need b) leading to enhancements of particular features that c) improve an organism's fit to its environment, which d) are then passed on and e) accumulate in each generation, f) leading to progressive increases in complexity, with g) no extinction, and h) simple forms produced anew by spontaneous generation. 4) Lamarck rejected the notion that the environment would directly affect organismal traits -- the point was that organisms responding to the environment led to adaptive changes that were passed on.
One person who postulated heritable, acquired, undirected variation was Darwin. ____ See also: The imaginary Lamarck by M. Ghiselin Shades of Lamarck by S.J. Gould Students' preconceptions about evolution: How accurate is the characterization as "Lamarckian" when considering the history of evolutionary thought? by K. Kampourakis and V. Zogza The early history of the idea of the inheritance of acquired characters and of pangenesis by C. Zirkle
|
| Functional redundancy. [Genomicron] Posted: 15 Jul 2008 03:15 PM CDT The move from Blogger to Scientific Blogging has been good overall, and I am certainly enjoying more hits and a broader readership from outside the normal blogosphere. However, I have also noticed that bloggers and blog readers have more or less dropped off. I think it would be useful for Genomicron to reach both audiences, but it seems that one Genomicron can't serve that function yet. Perhaps when the new format of SB is operational it will. Meanwhile, I am opting to resurrect the pseudoGenomicron and have it persist as a functionally redundant parablog. That is to say I will be cross posting everything on both sites, and blog readers who like this one better can read it and people who want links to lots of other interesting stuff can visit the other one. It will be a little more work, but worth it I think. The Feedburner feed will use the original Genomicron so it can have full posts. If you prefer the other version, it's here. |
| A Rundown of iPhone Health Apps [Epidemix] Posted: 15 Jul 2008 02:05 PM CDT Out of the bounty of new apps for the iPhone, I was pleased to see a couple dozen focused on health & fitness. To me, the potential here is this: Combine a device that’s easy to use and portable with the growing trend of life-logging. The result, I hope, would be several apps that let us track our health, quantatively, and log progress and data (basically, the idea would be not unlike the Virgin HealthMiles product I blogged about recently). So here’s my cursory rundown of the new iPhone apps that look most promising. Diet & Nutrition
Fitness
Medicine/Medical Records In addition to a couple pregnancy calculators (the Wheel for $14.99 and Birth Buddy for $4.99), these mostly fall in as health records tools - LifeRecord - or electronic emergency cards - EmergenKey ($1.99) ICE ($.99) and Emergency Card ($2.99). The personal health record tools look cool - you can store MRIs and other image files - but I wonder how compatible they are with Google Health or any other PHR out there (I imagine not at all). The emergency cards seem like a good idea, but fatally flawed: You really want to count of a EMT to find your iPhone, then click on the right app to find your health info? Seems unlikely to me. But two nifty apps here:
|
| Posted: 15 Jul 2008 01:37 PM CDT I think we have blogged about this crazy parasite previously. From SciencePunk: Once mature, S. tellinii needs to return to water to complete its life cycle. To do this, the Gordian worm infects the cricket's mind, and forces it to commit suicide. Little is known about how the worm achieves this, but French scientists recently reported that the expression of certain proteins in the infected cricket's brain were altered, namely those involved in neurotransmitter function and geotactic behaviour. The partly-digested and now zombified cricket seeks out water, and once there, throws itself in, and drowns. The adult Gordian worm then burrows out of the cricket carcass, swimming away to find a mate. So uninterested is the Gordian worm in the non-parasitic stage of its life cycle that it doesn't even have a mouth - it will live on the reserves built up whilst inside the host and then die. Each female Gordian worm can lay up to 10 million eggs. Bad news for crickets, good news for zombie fans. Here is a video that is making it's way through the intertubes, of the suicide and emergence from the cricket of this creepy parasite. |
| What’s on the web? (15 July 2008) [ScienceRoll] Posted: 15 Jul 2008 01:11 PM CDT
 |
| E.O. Wilson profile [Yann Klimentidis' Weblog] Posted: 15 Jul 2008 10:52 AM CDT ...in the New York Times, about ant species (14,001 of them!!!), recent paper on group selection, Dawkins' response in New Scientist (which I can't seem to locate), and sociobiology. see also John Hawks and Razib. |
| Pac Bio raises $100mill for sequencer commercialization [SEQanswers.com] Posted: 15 Jul 2008 10:12 AM CDT |
| Did You Eat Your Fruits and Vegetables Today? [Highlight HEALTH] Posted: 15 Jul 2008 09:30 AM CDT
The findings, published in the Nutrition Journal, demonstrate that self-reports of fruit and vegetable consumption are susceptible to substantial social approval bias [1]. Such biases are the presence of social desirability (the tendency to respond in such a way as to avoid criticism) and social approval (the tendency to seek praise) [2-3]. Researchers from the University of Colorado Denver randomly selected 163 women to complete what they were told would be a future telephone survey about health. Randomly half the women were sent a letter prior to the interview describing it as a study of fruit and vegetable intake. Included with the letter was a brief statement on the benefits of fruit and vegetable consumption, a 5-A-Day sticker and a 5-A-Day refrigerator magnet. The other half of the women in the study recieved the same letter but it described the study purpose only as a more general nutrition survey and contained neither the fruit and vegetable message nor the 5-A-Day materials. Within 10 days of receiving the letters, each of the women answered a food frequency questionnaire and were asked how many fruits and vegetables they had eaten in the last 24 hours. Since the two groups were randomly selected, fruit and vegetable consumption should have been the same in each group. However, those women that recieved the fruit and vegetable message and the 5-A-Day materials reported a significantly higher intake of total fruits and vegetables, and 40% of individuals were categorized as eating 5 or more servings of fruits and vegetables a day compared to just 18% in the other group. In response to the 24-hour recall questions, 61% of women who received the fruit and vegetable message and the 5-A-Day materials reported eating fruits and/or vegetables on three or more occasions throughout the previous day compared to 32% in the other group. These percentages were independent of age, race, educational level, self-perceived health status and time since last medical check-up. According to the authors of the study [1]:
This doesn’t mean that health questionnaires are useless. Rather, it means that many people simply aren’t being entirely truthful when it comes to how many fruits and vegetables they eat. The authors suggest that, in large dietary intervention trials, subgroups can be evaluated with biomarkers or other independent assessments to estimate the reporting bias size. Additionally, bias can also be controlled to some degree by assessing different intensities of an intervention, whereby everyone receives at least a minimal prompt for change instead of one group not receiving any prompt at all. A growing body of evidence shows that fruits and vegetables are critical to promoting good health. Here’s 5 tips to help you eat more:
Fruits and vegetables contain essential vitamins, minerals and fiber. In addition to the numerous health benefits of eating more fruits and vegetables, increased consumption lowers your risk of developing several cancers. Additional resources can be found in the Nutrition category of the Highlight HEALTH Web Directory. How about you? Do you lie to yourself about the fruits and vegetables you eat? References
.gif) Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership! Thank you for subscribing by RSS or email. I work hard to make the articles on Highlight HEALTH engaging and I truly appreciate your interest and readership!This article was published on Highlight HEALTH. Other Articles You May Like |
| Minority Report meets Gattaca! [The Gene Sherpa: Personalized Medicine and You] Posted: 15 Jul 2008 09:13 AM CDT |
| Science Tuesday: Back into the hornets’ nest [chrisdellavedova.com » Science] Posted: 15 Jul 2008 07:11 AM CDT
As you might expect, that’s not an easy question to answer. As is the case with most complex disorders there is no one “cause”. Autism isn’t like tuberculosis, there’s not a bacteria that causes the disease. In fact,most researchers believe that “autism” is not a discrete disorder, rather “autism is a clinically defined pervasive developmental disorder with phenotypically diverse neuropsychiatric symptoms and characteristics. These manifest as a spectrum of social and communicative deficits, stereotypical patterns and disturbances of behaviour.”¹ This spectrum of symptoms is collectively described as autism spectrum disorders (ASDs) and range from severe mental retardation often accompanied by seizures to milder . Symptoms of ASDs usually become apparent in children by three years old, but are often detectable by 14 months. This is one of the reasons that the unsubstantiated ‘link’ between autism and the MMR vaccine is so persistent, the age at which the vaccine is given is often the age at which symptoms become apparent. There has been extensive research into the neuropathology and neurobiology of ASDs and a number of changes to the nervous system are associated with autism. These include aberrations in brain growth, neuronal patterning and cortical connectivity as well as changes in structure and function of synapses and dendrites.² The cause of these morphological changes, and thus autism, is not so clear. The short answer is ASDs are primarily genetic disorders, but there may be environmental factors that are involved as well. For the long answer, read on. Genes: This is the big one. In the case of autism, as with many complex traits, the heritability of the trait can be estimated. Heritability refers to the proportion of phenotypic variation that is attributable to genetic variation. If a particular trait’s heritability is 100% then the trait is due entirely to genetic variation, if the heritability is 0% then the trait is due entirely to environmental variation. By some estimates, heritability of autism spectrum disorders exceeds 90%. Twin studies support a strong genetic component and sibling recurrence risk - the chance of a younger sibling of an autistic child having autism, exceeds 15%. When compared with the rates in the general population (1 in 500 for “autism” and 1 in 150 for ASDs) there is little doubt that a big piece of the puzzle rests in our genes.³
Walsh’s group found a number of different mutations in autistic children from these families, a number of which were associated with large deletions of parts of the genome or chromosomal rearrangements. A subset of these could be associated with mutations in autistic children with unrelated parents, lending support to the hypotheses that defects in these genes cause ASDs. Interestingly, some of the genes identified by Walsh’s group and others are also associated with other neurological disorders including Fragile X syndrome, Rett syndrome and epilepsy. As a geneticist, it is a temptation to end this post here. To declare that the cause of autism is a series of mutations in the genome that induce changes in brain morphology and the spectrum of symptoms that we call autism. Unfortunately, that would be ignoring part of the equation. Some researchers think that the 90% heritability estimate is high, and even if it is not the environment still plays a role in autism. Environmental factors: There has been a wealth of research into the role of environmental toxins in causing autism and this school of thought is the one that supports the MMR-autism link. The argument here is that the massive increase in autism in the last couple of decades points to an environmental influence for which genetics alone can not account. Researchers are looking most closely at the usual suspects - heavy metals like arsenic, lead and mercury. There is some evidence to support an association between even low level exposure to toxic heavy metals and neurological issues ADHD and lower IQs. This association led to the scare regarding the mercury containig thimerosal in the MMR vaccines. However, repeated studies have found that autism diagnoses continue to rise even after the removal of thimerosal from the vaccine.
Finally, when thinking about the enviromental influences on autism, it’s important to explore the role of the environment on genetics. Many of the types of genetic changes that have been identified as causative in autism are indicative of some sort of DNA damage - DNA damage that may result from exposure to an environmental toxin. Many scientists, and I count myself in their number, feel that the recent autism ‘epidemic’ is due primarily to improved screening and diagnosis. In other words, prior to the 1980’s, many people suffering from autism were diagnosed as “slow” or misdiagnosed with another type of mental retardation. Unfortunately, there is no way to quantify this hypothesis. The alternative is that there is one or more environmental components that we increasingly exposed to that is causing more frequent incidents of autism. Perhaps these environmental factors are acting as a DNA mutagen - a compound that causes DNA changes. If that is the case, we must consider timing. Autistic children bearing causative mutations were born with those mutations. They originated either de novo at a very early stage of development, possibly as a result of exposure to some toxin in utero, or they came from the parents. The take home message is that researchers do not know what causes autism in children. Most evidence supports that genetics plays the dominant role, but if in fact the frequency of autism is rising, then there are very likely some compounds in our environment that play a role. My Ph.D. supervisor always told me that as scientists we can never “prove” the truth, only disprove falsehoods. That’s where we are with the causes of autism. Researchers have disproved the falsehood that the MMR vaccine causes autism. However, they can’t assuage parents anxieties by collaring the culprit. ——————- 1: Schmitz and Rezaie. The neuropathology of autism: where do we stand? Neuropathology and Applied Neurobiology 2008; 34: 4 -11 2: Pardo and Eberhart. The Neurobiology of Autism. Brain Pathology 2007; 17: 437 -447. 3: Sutcliffe. “Insights into the Pathogenesis of Autism. Science 2008; 321: 208-9. ——————- The Hold Steady’s - “Separation Sunday” is available from This posting includes an audio/video/photo media file: Download Now |
| Resistance is Futile [The Gene Sherpa: Personalized Medicine and You] Posted: 15 Jul 2008 06:47 AM CDT |
| The Price of PBS These Days! [Bitesize Bio] Posted: 15 Jul 2008 06:28 AM CDT It seems that everyone is throwing their hands up about gas/petrol prices these days. In the US, gas prices have now reached $4/gallon, while in the UK, they are hovering around a whopping $10/gallon. But what’s all the fuss about? A lot of effort goes into producing gas - you have to find, extract, refine, transport, and of course, tax it (especially in the UK) none of which is cheap. So maybe gas consumers are getting value for money. Well, compared to lab workers they certainly seem to be. For some of the liquids available for molecular/cell biology, some of which are much easier to produce than petrol, $4 or even $10/gallon is peanuts. Here are some examples to help keep things in perspective next time you are at the pump. Distilled water: Run the tap, pass the water through a distillation unit, then maybe through a sub-micron filter to clean it up at the end, bottle, then ship. A bargain at $75/gallon. PBS: A salty version of distilled water. Must be expensive salt because it raises the price to $100/gallon. 10x TBE: Slightly more elaborate salty distilled water, coming in at $150/gallon Molecular biology grade water: In the lab, we used to call this holy water because many people swear by it. It’s just very clean water, but how much cleaning can you do for $300/gallon? DEPC treated water: The price of being RNAse free? $350/gallon, apparently. Dulbecco’s Modified Eagle Medium: Cell food. £375/gallon LB Medium: Water, salt, Marmite and chopped up soya beans. $400/gallon Ethidium bromide: The stuff of the devil according to some (but not all), the price is devilish though… $25,000/gallon EcoRI: To be fair, this stuff takes a lot of effort to make but, just for fun, a standard prep comes in at $375,000/gallon All food for thought so maybe we should lay off those poor impoverished oil companies. Or maybe not. What’s the most over-priced thing you’ve bought for the lab? Photo: lorenzodom |
| MIT-led team finds language without numbers [Think Gene] Posted: 15 Jul 2008 04:41 AM CDT Kevin: This builds on earlier studies about the Piraha showing they have the numbers zero, one and many. An Amazonian language with only 300 speakers has no word to express the concept of “one” or any other specific number, according to a new study from an MIT-led team. The team, led by MIT professor of brain and cognitive sciences Edward Gibson, found that members of the Piraha tribe in remote northwestern Brazil use language to express relative quantities such as “some” and “more,” but not precise numbers. It is often assumed that counting is an innate part of human cognition, said Gibson, “but here is a group that does not count. They could learn, but it’s not useful in their culture, so they’ve never picked it up.” The study, which appeared in the June 10 online edition of the journal Cognition, offers evidence that number words are a concept invented by human cultures as they are needed, and not an inherent part of language, Gibson said. The work builds on a study published in 2004, which found that the Piraha had words to express the quantities “one,” “two,” and “many.” The MIT researchers observed the same phenomenon when they asked Piraha speakers to describe sets of objects as they were added, from one to 10. However, the MIT team decided to add a new twist–they started with 10 objects and asked the tribe members to count down. In that experiment, the tribe members used the word previously thought to mean “two” when as many as five or six objects were present, and they used the word for “one” for any quantity between one and four. This indicates that “these aren’t counting numbers at all,” said Gibson. “They’re signifying relative quantities.” He said this type of counting strategy has never been observed before, although it may also be found in other languages believed to have “one,” “two,” and “many” counting words. The paper is part of a larger project that investigates the relationship between Piraha culture and their cognition and language, testing some claims by Daniel Everett, a linguist at Illinois State University, in a 2005 issue of Current Anthropology. One other discovery of the project is that the Piraha can perform exact matching tasks as long as there is no memory component to them, but once there is a memory component, they approximate their matches. This suggests that language is a cognitive technology that aids humans in memory tasks. Lead author of the paper is Michael Frank, a graduate student in Gibson’s lab. Other authors are Evelina Fedorenko, a postdoctoral associate at the McGovern Institute for Brain Research at MIT, and Everett. Source: Anne Trafton, MIT News Office |
| The billion-dollar question: is big science worth the money? [Genetic Future] Posted: 15 Jul 2008 03:06 AM CDT Dan Koboldt from MassGenomics has a great article on the Cancer Genome Atlas (TCGA) project, an ambitious and seriously expensive endeavour to catalogue the genetic changes that occur during the transition of well-behaved human cells into the sprawling, anarchic lethality of cancer. How expensive? Well, the pilot project cost $100 million, so this is going to be a costly effort even by genomics standards; the total expense, over the next ten years or so, may reach $1.5 billion according to a recent news article in the journal Science. Dan uses the project as a springboard to weigh up the merits of the Big Science approach - research performed on a massive scale by large, multi-centre collaborations rather than the traditional small-scale, more focused work performed by individual labs. The benefits of the Big Science approach are obviously embodied in the public Human Genome and HapMap projects, both of which have utterly changed the way human genetics is done. In fact, it's hard to think of a recent Big Science project in human genetics that hasn't exceeded expectations, to the horror of its inevitable early detractors. Dan hints that Evan Eichler's recent $40 million grant to categorise structural variation in the human genome may be an example of Big Science money that could have been more effectively spent on other projects, but it's probably too soon to judge this with any certainty. And while it's true that the recent rash of genome-wide association collaborations such as the Wellcome Trust Case Control Consortium have captured a disappointingly small (indeed, sometimes non-existent) fraction of the genetic risk for many common diseases, they have also provided powerful insight into the genetic architecture of human traits, novel information about disease mechanisms, and a set of massive cohorts with high-quality DNA samples to drive future large-scale sequencing projects. Perhaps the biggest danger of the phenomenal success of the Human Genome and HapMap projects is that this success provides a fully general counter-argument to wield against nay-sayers for any future Big Science endeavour, however foolish. ("Oh, so you claim that my proposed project to sequence the human [whatever]ome is too expensive and technically impossible? Well, they said the same thing about the HapMap project!" Opponents exit, broken, stage right; funding agencies enter with bags of cash, stage left.) |
| Next-next-gen sequencer raises $100M [Genetic Future] Posted: 15 Jul 2008 02:08 AM CDT  As I've mentioned a couple of times here, the current crop of personal genomics products (which scan a meagre 500,000 to a million DNA positions - a fraction of 1% of your complete genome) are basically place-holders for the rapidly approaching era of affordable whole-genome sequencing. As I've mentioned a couple of times here, the current crop of personal genomics products (which scan a meagre 500,000 to a million DNA positions - a fraction of 1% of your complete genome) are basically place-holders for the rapidly approaching era of affordable whole-genome sequencing.At the moment the rapid sequencing market is being dominated by three "next-generation" sequencing platforms: Roche's 454, Illumina's Solexa, and the SOLiD platform from Applied Biosystems (which was recently gobbled up by consumables giant Invitrogen). These three platforms are currently enthusiastically jostling for position - for instance, competing with each other to offer their services to the international 1000 Genomes Project - in an attempt to establish themselves as the technology of choice as the age of medical genomics looms ever closer. With each evolutionary iteration of these technologies their sequencing capacity and accuracy improves, enabling truly mind-boggling amounts of sequence to be generated (exhibit A: the Sanger Institute's recent one trillion base pair milestone). But while these next-gen technologies - each with hefty financial backing from a major company - are stacking up the bases, a new generation of upstart "next-next-generation" platforms is developing their own revolutionary approaches to sequencing. One of the most promising of these platforms is the SMRT system from Pacific Biosciences, which uses fictional-sounding technology called zero-mode waveguides to detect the incorporation of fluorescent bases into a single, immobilised DNA molecule (you can see for yourself how the PacBio system works in this pretty promotional video). Apparently PacBio's technology (along with the fancy graphics, and the fact that it has zero-mode waveguides!) has convinced the people that matter: GenomeWeb News notes that the company has raised a hefty $100 million from investors to develop a commercial platform, which is expected to hit the market in 2010. In other words, the already-seething large-scale sequencing market will soon be getting even more interesting. As I've noted before, I really don't care who wins this technological arms race: all that matters to me is that so long as it's being run my own genome sequence (and yours) is becoming rapidly more affordable. (Image from Pacific Biosciences.) |
| The Problem is Too Much Biological Complexity [adaptivecomplexity's column] Posted: 14 Jul 2008 09:20 PM CDT We like to talk about the amazingly complex machinery of the cell: flagella that resemble finely tuned outboard motors, or complex information processing circuits that help a cell process information about its environment. Biologists work hard at understanding how these systems work. They will take a wiring diagram like the following, and ask why is it set up this way? |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |


 of his theory of natural selection to explain the evolution of species. He spent the next 20 years developing the theory and marshaling evidence in favour of both the fact that species are related through common descent and his particular theory to explain this. After receiving word that another naturalist,
of his theory of natural selection to explain the evolution of species. He spent the next 20 years developing the theory and marshaling evidence in favour of both the fact that species are related through common descent and his particular theory to explain this. After receiving word that another naturalist,  It is a general law of nature for all creatures to propagate the like of themselves: and this extends even to the most trivial minutiæ, to the slightest individual peculiarities; and thus, among ourselves, we see a family likeness transmitted from generation to generation. When two animals are matched together, each remarkable for a certain given peculiarity, no matter how trivial, there is also a decided tendency in nature for that peculiarity to increase; and if the produce of these animals be set apart, and only those in which the same peculiarity is most apparent, be selected to breed from, the next generation will possess it in a still more remarkable degree; and so on, till at length the variety I designate a breed, is formed, which may be very unlike the original type.
It is a general law of nature for all creatures to propagate the like of themselves: and this extends even to the most trivial minutiæ, to the slightest individual peculiarities; and thus, among ourselves, we see a family likeness transmitted from generation to generation. When two animals are matched together, each remarkable for a certain given peculiarity, no matter how trivial, there is also a decided tendency in nature for that peculiarity to increase; and if the produce of these animals be set apart, and only those in which the same peculiarity is most apparent, be selected to breed from, the next generation will possess it in a still more remarkable degree; and so on, till at length the variety I designate a breed, is formed, which may be very unlike the original type. 
 ...if an organised body is not in the situation and circumstances best adapted to its sustenance and propagation, then, in conceiving an indefinite variety among the individuals of that species, we must be assured, that, on the one hand, those which depart most from the best adapted constitution, will be most liable to perish, while, on the other hand, those organised bodies, which most approach to the best constitution for the present circumstances, will be best adapted to continue, in preserving themselves and multiplying the individuals of their race.
...if an organised body is not in the situation and circumstances best adapted to its sustenance and propagation, then, in conceiving an indefinite variety among the individuals of that species, we must be assured, that, on the one hand, those which depart most from the best adapted constitution, will be most liable to perish, while, on the other hand, those organised bodies, which most approach to the best constitution for the present circumstances, will be best adapted to continue, in preserving themselves and multiplying the individuals of their race.  Half a century before Charles Darwin published On the Origin of Species, the French naturalist Jean-Baptiste Lamarck outlined his own theory of evolution. A cornerstone of this was the idea that characteristics acquired during an individual's lifetime can be passed on to their offspring. In its day, Lamarck's theory was generally ignored or lampooned. Then came Darwin, and Gregor Mendel's discovery of genetics. In recent years, ideas along the lines of Richard Dawkins's concept of the "selfish gene" have come to dominate discussions about heritability, and with the exception of a brief surge of interest in the late 19th and early 20th centuries, "Lamarckism" has long been consigned to the theory junkyard.
Half a century before Charles Darwin published On the Origin of Species, the French naturalist Jean-Baptiste Lamarck outlined his own theory of evolution. A cornerstone of this was the idea that characteristics acquired during an individual's lifetime can be passed on to their offspring. In its day, Lamarck's theory was generally ignored or lampooned. Then came Darwin, and Gregor Mendel's discovery of genetics. In recent years, ideas along the lines of Richard Dawkins's concept of the "selfish gene" have come to dominate discussions about heritability, and with the exception of a brief surge of interest in the late 19th and early 20th centuries, "Lamarckism" has long been consigned to the theory junkyard.



 Call me a glutton for punishment, but despite
Call me a glutton for punishment, but despite  The problem is that autism is not caused by a mutation in a single gene. Nearly 30 individual genes have been identified as playing a role in ASDs, including genes involved in synapse function, neuron adhesion, endosomal trafficking, neuronal activity regulation and other biological processes. The
The problem is that autism is not caused by a mutation in a single gene. Nearly 30 individual genes have been identified as playing a role in ASDs, including genes involved in synapse function, neuron adhesion, endosomal trafficking, neuronal activity regulation and other biological processes. The  A second interesting line of research looks at a different type of environmental influence - maternal viral infections in autism in children. Since the mid-90’s researchers have known that a strong correlation exists between maternal influenza infection, particularly in the second trimester, and the likelihood of having autistic children. Recent research has involved attempting to discern the reason for this phenomena by using animal models. A recent paper describes a mouse system in which adult progeny of virally infected mothers display a number of ASD symptoms. Beyond the potential causative effects of virus infection, a mouse model would be of great utility for neuropathology, epidemiology and potentially development of treatments for autism.
A second interesting line of research looks at a different type of environmental influence - maternal viral infections in autism in children. Since the mid-90’s researchers have known that a strong correlation exists between maternal influenza infection, particularly in the second trimester, and the likelihood of having autistic children. Recent research has involved attempting to discern the reason for this phenomena by using animal models. A recent paper describes a mouse system in which adult progeny of virally infected mothers display a number of ASD symptoms. Beyond the potential causative effects of virus infection, a mouse model would be of great utility for neuropathology, epidemiology and potentially development of treatments for autism.
No comments:
Post a Comment