The DNA Network |
| Google launches new Web browser [Microarray and bioinformatics] Posted: 02 Sep 2008 03:22 PM CDT Google has launched the new google browser in Beta version. The Google Chrome (BETA) for Windows is claiming to be a browser that combines a minimal design with sophisticated technology to make the web faster, safer, and easier. I downloaded it and is making this post fom the new browser , its fast is my first comment, but it still uses the IE engine just like Fiefox for its security features Have you heard of the new feature in Internet Explorer 8 called InPrivate Browsing or rather nicknamed as Pron Mode, Macintosh users already know about this features that deletes the track record of your action in the Browser. Chomr also has a similar feature called incognito mode. Am not that impressed I just ask my firefox to delete the cache and history automoatically every time I close the browser. For More Details take a look at the website  |
| Posted: 02 Sep 2008 02:15 PM CDT On my latest Calgary hunt for Biopunk material, I was curious as to whether various book and comic specialty shop clerks had any Biopunk books. I was surprised to hear that most stores didn’t have a clue as to what the term meant, nor really showed an interest other than the obligatory computer search. I had gone to a few bookstores and comic stores before ending my day at The Sentry Box. I was overjoyed to see that the staff was absolutely passionate about Sci-fi and Gaming, even offering to watch my toddler while I looked around. They were amazingly helpful and interested when I asked if they had anything Biopunk. They hadn’t heard of the term either. I suppose that’s due to the relative newness of the genre, but once I explained they gleefully directed me to a number of novels and a book called GURPS Bio-Tech. The GURPS fourth edition (2006) “Bio-Tech” sourcebook by David Morgan-Mar and David Pulver is a different medium for those of you who are might like to mix your Biopunk and Role Playing Games (RPGs). This is also an excellent inspiration and resource for aspiring Biopunk writers. |
| Large scale phasing and imputing in Iceland [Mailund on the Internet] Posted: 02 Sep 2008 12:31 PM CDT There is a really cool paper in the latest issue of Nature Genetics by people from deCODE:
As the abstract says, it concerns haplotype phasing and imputation, but the setup is really cool! The case of IcelandIceland is a bit special. The Icelandic population is relatively small (about 300,000) and about 10% of the population has been “genome wide” genotyped at deCODE. This is a very large fraction of the population, by any standard. Further, the pedigree of the population is fairly well know from historical records and estimated to be both reasonably complete and reasonably accurate for the last few centuries. Again, this is rather unique. Now, this paper introduces a method that exploits these two facts to both impute haplotype phase and impute genotype information for untyped individuals (yes, individuals, not just missing markers!) Trios and trio proxiesInferring the haplotype phase of an individual is much simplified if you know the genotypes of his parents. For a parent-child trio, the homozygotic sites in the parents can be used to infer the phase of the heterozygotic sites in the child. If the child is heterozygotic Aa but the father is homozygotic AA, then clearly the A allele comes from the father. This simple observation can be used to infer haplotype phase. It won’t resolve all sites, of course, since it doesn’t help anything at sites heterozygotic in all three, but it does resolve a lot of sites. Now, typically you do not have trios in an association mapping study. Population based association mapping studies requires to a large degree that the individuals are unrelated, so you would only be able to use the parents anyway, and those are not the ones you can phase this way. |
| The lighter side of giving a lecture [Discovering Biology in a Digital World] Posted: 02 Sep 2008 12:17 PM CDT David Ng from The World's Fair has made this wonderful video on public speaking. Or shall we say the funnier side of public speaking? I really enjoyed it! I'd say Dave's video ranks right up there with one of my other favories, the classic: "Chicken chicken chicken"
|
| Innovation Slow-Down in the US? [adaptivecomplexity's column] Posted: 02 Sep 2008 11:18 AM CDT The technology community in the US ain't what it used to be, argues Judy Estrin, a technology entrepreneur and former Chief Technology Officer of Cisco. She is the author of Closing the Innovation Gap, and she's arguing that "we have a national innovation deficit." |
| European genetic structure [Yann Klimentidis' Weblog] Posted: 02 Sep 2008 11:02 AM CDT There's been two recent papers that look at the relationship between geography/ethnicity and genetics in Europe. Dienekes has some excellent posts and pictures of these two papers (here, here), as well as at GNXP (here, here, here). from the Nature paper: an individual's DNA can be used to infer their geographic origin with surprising accuracy--often to within a few hundred kilometres.and from a news story: The map was so accurate that when Novembre's team placed a geopolitical map over their genetic "map", half of the genomes landed within 310 kilometres of their country of origin, while 90% fell within 700 km. |
| Let's talk about the facts this election - Obama on Science Funding [The Daily Transcript] Posted: 02 Sep 2008 09:24 AM CDT This weekend I moved from the Backbay to Cambridgeport, aka Junior Faculty ghetto. It's conveniently located at the near BU, Harvard and MIT. It is probably one of the most overeducated (if such a term existed) neighborhoods on the planet. So, in today's "let's talk about the facts" entry I'll encorage you to check out JuniorProf's entry. Read the comments on this post... |
| Constellation Pharmaceuticals Aims to Develop Epigenetic Drugs [Epigenetics News] Posted: 02 Sep 2008 09:01 AM CDT One of the news items that was missed in April during the brief hiatus was about a new biotech outfit (Constellation Pharmaceuticals) that had raised $32 million in a series A round of funding. The company immediately drew my attention not because they were planning to develop epigenetic drug therapies for cancer, but because of their founders: David Allis, Yang Shi, and Danny Reinberg are all well-respected scientists in the epigenetics field, and their founding of the company brings an immediate legitimacy to the organization, which was no doubt helpful in the round of funding. Additionally, it probably didn’t hurt gaining the attention of Mark Levin of Third Rock Ventures, the former cofounder of Millenium Pharmaceuticals, who is now the interim CEO of Constellation. The young company’s web site is full of promising publicity talk about the potential of epigenetic therapies, but little of material value. However, the company is quite young and it will likely take awhile before any promising candidates emerge. As a side note, if you’re a BS/MS-level graduate in molecular biology looking for work, Constellation is hiring in the Boston area. For more information about Constellation, see this article from the Boston Globe. |
| Genetic Future is moving, and so am I [Genetic Future] Posted: 02 Sep 2008 08:20 AM CDT Genetic Future is moving to a shiny new home at ScienceBlogs. This domain will remain as an archive site, but for fresh content you will need to update your links as follows: New URL: http://scienceblogs.com/geneticfuture/ New RSS feed: http://feeds.feedburner.com/scienceblogs/geneticfuture Some of you familiar with the ScienceBlogs network might be wondering if this move heralds a transition into left-wing political blogging, but don't worry: my articles will continue to be focused on reporting advances in human genomics and critiquing the genetic testing industry. Just a few weeks after the transition I'll also be physically moving from Sydney to a new life in Cambridge, UK. Posting on the new site will be light during this move and regulars will notice a few recycled posts to fill in the awkward silences, but bear with me - in a couple of weeks there will be plenty of fresh human genetics goodness. Hope to see you all at the new domain, Daniel. |
| Posted: 02 Sep 2008 07:15 AM CDT Busy preparing for the start of the semester, so to tide you over here are some links of things to check out. 1) In our genes, old fossils take on new roles by David Brown, Washington Post It turns out that about 8 percent of the human genome is made up of viruses that once attacked our ancestors. The viruses lost. What remains are the molecular equivalents of mounted trophies, insects preserved in genomic amber, DNA fossils. 2) Gaming evolves by Carl Zimmer, New York Times
I may have to check out this game. 3) Research raises questions about DNA barcoding methodology "Sadly, the authors of this paper do not understand barcoding protocols," Paul Hebert, director of the Biodiversity Institute of Ontario at the University of Guelph, told GenomeWeb Daily News. Calling the title of the paper misleading, he said barcoders have been aware of nuclear pseudogenes for years and have already designed some strategies for dealing with the problems described in the paper. |
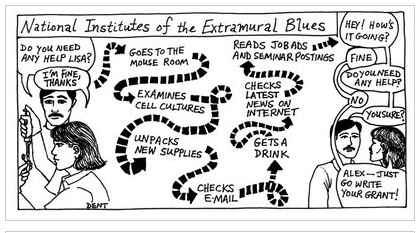
| Dent Cartoon Archive [Bitesize Bio] Posted: 02 Sep 2008 05:50 AM CDT Alex Dent’s hilarious cartoons that caricatured the turmoil of life as a postdoc are a must see. Alex produced most of his cartoons during early 90’s, and lit up many an edition of the NIH Catalyst newsletter, until he finally gave up cartooning in 2002. But the good news is that earlier this year, with a little help from his friends, Alex set up an archive of cartoons so that future generations can understand the 9 types of principle investigators, sing the post-doc blues and be fore-warned about the three stages of Post-doc life. Check them all out at dentcartoons.blogspot.com. |
| Google Chrome [Mailund on the Internet] Posted: 02 Sep 2008 05:05 AM CDT I was just told about this post. Apparently, Google is developing its own browser. It is described in this cartoon that features people I know here from Google in Aarhus such as Lars Bak and Kasper Verdich. I didn’t know what they were working on. A lot of them were working on virtual machines for mobile phones and such before going Google, so I thought it was something like, but apparently not. It’s still virtual machines, though, from what I get from the cartoon. |
| wellcome to DNA network [Reportergene] Posted: 02 Sep 2008 03:24 AM CDT  In order to better reach that hidden niche of passionates, Reportergene will proudly broadcast its specialized view in reportergenomics to the DNA network, a group of up to 50 entusiast bloggers joined together in a feedburner network. If you're looking for information regarding DNA, genes, genomes and (of course) any reporter gene, subscribe the DNA Network RSS feed that aggregates all these amazing blogs into one power packed feed source. In order to better reach that hidden niche of passionates, Reportergene will proudly broadcast its specialized view in reportergenomics to the DNA network, a group of up to 50 entusiast bloggers joined together in a feedburner network. If you're looking for information regarding DNA, genes, genomes and (of course) any reporter gene, subscribe the DNA Network RSS feed that aggregates all these amazing blogs into one power packed feed source.Read older site news |
| Genetics, Evo-devo and Spore [business|bytes|genes|molecules] Posted: 02 Sep 2008 01:15 AM CDT
What are the things that evolution has at its disposal to define a creature, to mix and match the parts, and eventually come up with a unique organism that's going to live its life and try to reproduce? Every time I take a “how big a geek are you” test, I fail, since I am not a gamer. That’s by choice. If I ever start, I am doomed. There is not enough time for all the things I care about as it stands, especially music, so gaming has always been kept at arms length. That said, cool games, and ideas in games are always of interest. Spore, which gets released on Sunday, has to be among the most highly anticipated, most hyped games since, well I suppose, GTA IV. Well, when you bring a cool game and genetics together, you get my attention. For those who have been living inside a cave, Spore allows players to design a virtual world of new life and then control the evolution of the creatures and species they create. Being a game, you aren’t exactly limited to life as we know it. You can create all kinds of creatures including such creatures as a one-eyed web-footed creature with a snout (I am sure you can get much more crazy than that). But apparently, the rules governing creature creation aren’t as random as one might think. On September 9th, the National Geographic Channel is premiering a documentary entitled How to build a better being. The documentary goes into the backstory of how Will Wright, the creator of Spore, worked with scientists like Michael Levine and Neil Shubin to understand the principles of genetics, mutations, evolution, biodiversity, etc. Since the game allows you to mate, compete against predators, obtain better body parts and, if they survive, ultimately become spacefaring voyagers, I suspect it will be interesting to see what the in game experience is like, and if the game can be used, perhaps in a modified form, or as controlled game play, to explain some of the underlying principles of evolution and genetics to kids. I have said many times that in a world where kids are exposed to interactive multimedia environments from a very young age, we need to make teaching interesting. Nat Torkington achieved that by using Scratch to teach kids programming. Since Spore is even cooler that Scratch, maybe we can leverage interest in Spore to teach children about how species evolved, what they share in common, the competitive pressures of nature, etc. For example, Neil Shubin was able to explain to Wright what the fundamental design patterns of animal life on earth are and was able to recreate a prehistoric fish in Spore. Here’s a video that was made available that shows some of the discussions mentioned above. It’s kinda cool to see the kind of discussion and planning that goes into a video game and you get the idea of how you might be able to use some of those ideas and implement them into a class curriculum, one that actually be fun. Related articles by Zemanta
|
| To Describe a Human [Think Gene] Posted: 31 Aug 2008 02:19 PM CDT Slobodan Cekic writes in response to How Much Data is a Human Genome:
740MB is the size of a reference human haploid nucleotide base string, not the data necessary to describe a mature human. We believe that most of our hereditary information resides in genes because it does. However, a genome, as you say, cannot possibly fully describe a mature human. A genome is more like a brief mathematical equation used to produce beautifully complex fractal design when fed with ambient noise and interpreted as colors and coordinates on a screen. Arguably, as other commentators have noted, this isn’t enough to describe a genome. Cytosine can be methylated —like a fifth base. Sometimes, a sequencing machine is unable to assert a base, and an extra bit would necessary to report these “no calls.” But in reality, humans don’t have a “reference genome.” Almost every cell has its own pair of genomes, and these tend to diverge as they accumulate mutations and errors. To be pedantic, to record your Real and Complete Human Genome, one would have to sequence every strand of DNA in your body instantaneously. Yet, these trillions of strings of millions of bases can be understood as that 740MB reference human genome like a field full of flowers can be understood as a photo of daisy. |
| You are subscribed to email updates from The DNA Network To stop receiving these emails, you may unsubscribe now. | Email Delivery powered by FeedBurner |
| Inbox too full? | |
| If you prefer to unsubscribe via postal mail, write to: The DNA Network, c/o FeedBurner, 20 W Kinzie, 9th Floor, Chicago IL USA 60610 | |



![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_b.png?x-id=b65b2e9c-985d-4ddd-93ff-5282d61ed1dd)

No comments:
Post a Comment